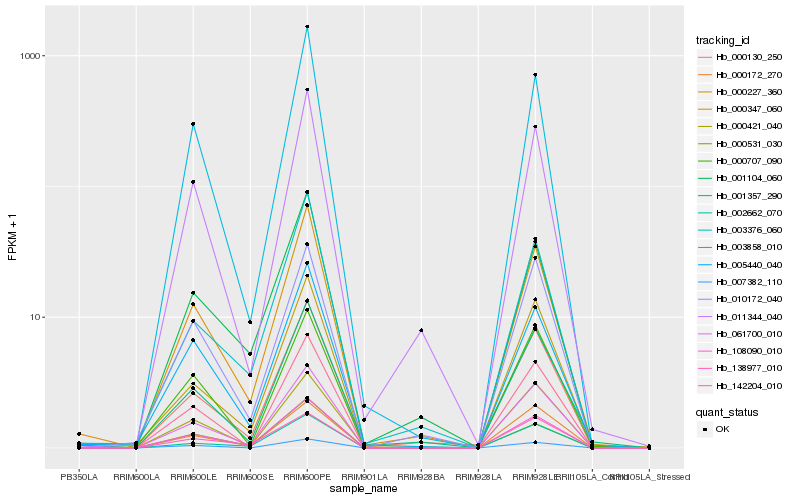
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108090_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_061700_010 |
0.0515865515 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 3 |
Hb_000227_360 |
0.0612165306 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 4 |
Hb_003858_010 |
0.0727246924 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 5 |
Hb_005440_040 |
0.0760815302 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 6 |
Hb_000130_250 |
0.0833212731 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |
| 7 |
Hb_000531_030 |
0.0854554343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002662_070 |
0.0861773124 |
- |
- |
hypothetical protein JCGZ_16102 [Jatropha curcas] |
| 9 |
Hb_001104_060 |
0.0869882024 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 10 |
Hb_011344_040 |
0.0872041816 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 11 |
Hb_142204_010 |
0.0883942999 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 12 |
Hb_010172_040 |
0.0896902094 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000421_040 |
0.0897159439 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 14 |
Hb_007382_110 |
0.0969450431 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 15 |
Hb_000172_270 |
0.0971759804 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 16 |
Hb_000707_090 |
0.0978371683 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 17 |
Hb_138977_010 |
0.0991679079 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_003376_060 |
0.0992595793 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000347_060 |
0.1006277995 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 20 |
Hb_001357_290 |
0.1028108307 |
- |
- |
PREDICTED: uncharacterized protein LOC105632986 [Jatropha curcas] |