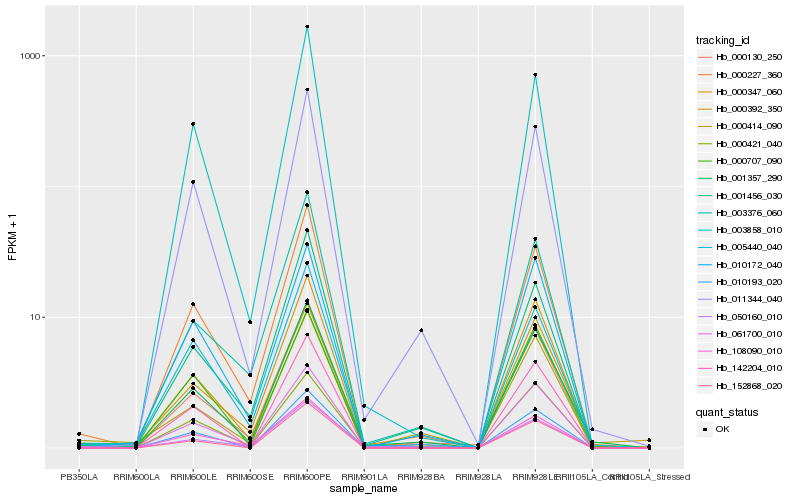
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632986 [Jatropha curcas] |
| 2 |
Hb_000227_360 |
0.0715262972 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 3 |
Hb_011344_040 |
0.0729595039 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 4 |
Hb_000130_250 |
0.0758899246 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |
| 5 |
Hb_000347_060 |
0.0835698873 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 6 |
Hb_061700_010 |
0.0885362186 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 7 |
Hb_000392_350 |
0.094796104 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_142204_010 |
0.0987653095 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 9 |
Hb_050160_010 |
0.099655467 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 10 |
Hb_108090_010 |
0.1028108307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001456_030 |
0.1039314936 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 12 |
Hb_152868_020 |
0.1039679975 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 13 |
Hb_000421_040 |
0.1049276466 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 14 |
Hb_010172_040 |
0.1053115812 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003858_010 |
0.1054251612 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 16 |
Hb_000414_090 |
0.1062600892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005440_040 |
0.1069129658 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 18 |
Hb_010193_020 |
0.1074469268 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 19 |
Hb_003376_060 |
0.1078500975 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000707_090 |
0.1078809673 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |