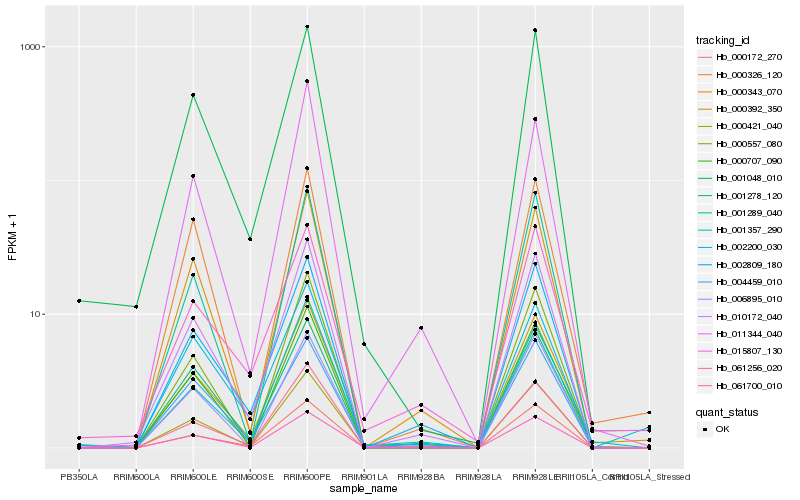
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_350 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000421_040 |
0.0811384988 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 3 |
Hb_010172_040 |
0.0816328418 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000707_090 |
0.0877870789 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 5 |
Hb_061700_010 |
0.0943820496 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 6 |
Hb_001357_290 |
0.094796104 |
- |
- |
PREDICTED: uncharacterized protein LOC105632986 [Jatropha curcas] |
| 7 |
Hb_002809_180 |
0.0949410936 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 8 |
Hb_000172_270 |
0.0953673641 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 9 |
Hb_000343_070 |
0.0956405699 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 10 |
Hb_061256_020 |
0.0979068721 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_000326_120 |
0.1020897838 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_006895_010 |
0.102611572 |
- |
- |
PREDICTED: secologanin synthase-like [Citrus sinensis] |
| 13 |
Hb_015807_130 |
0.103362587 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000557_080 |
0.1036136638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001289_040 |
0.1042696367 |
- |
- |
NDR1/HIN1-like 1 [Theobroma cacao] |
| 16 |
Hb_001278_120 |
0.1047923773 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 17 |
Hb_011344_040 |
0.1065432457 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 18 |
Hb_001048_010 |
0.107760659 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |
| 19 |
Hb_002200_030 |
0.1080942078 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 20 |
Hb_004459_010 |
0.1088815747 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |