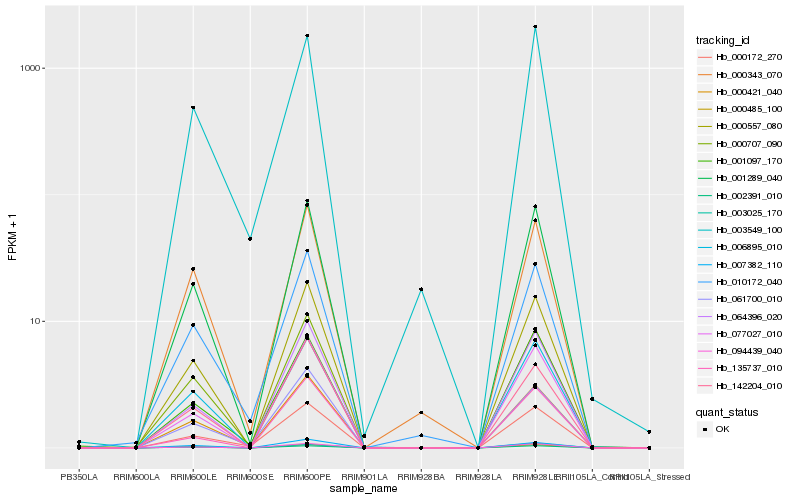
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001289_040 |
0.0 |
- |
- |
NDR1/HIN1-like 1 [Theobroma cacao] |
| 2 |
Hb_094439_040 |
0.0188077676 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 3 |
Hb_000557_080 |
0.0379952966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_077027_010 |
0.0539233405 |
- |
- |
hypothetical protein JCGZ_04511 [Jatropha curcas] |
| 5 |
Hb_006895_010 |
0.0542088826 |
- |
- |
PREDICTED: secologanin synthase-like [Citrus sinensis] |
| 6 |
Hb_064396_020 |
0.0571312113 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 7 |
Hb_000421_040 |
0.0612127968 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 8 |
Hb_000707_090 |
0.0627202629 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 9 |
Hb_001097_170 |
0.070835449 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 10 |
Hb_000172_270 |
0.0720084789 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 11 |
Hb_000485_100 |
0.0810691764 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 12 |
Hb_061700_010 |
0.0836314819 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 13 |
Hb_010172_040 |
0.0850647513 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003025_170 |
0.0878205571 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |
| 15 |
Hb_000343_070 |
0.0898139074 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 16 |
Hb_142204_010 |
0.0942007399 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 17 |
Hb_003549_100 |
0.0953976595 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 18 |
Hb_007382_110 |
0.0965767479 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 19 |
Hb_002391_010 |
0.0974162479 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 20 |
Hb_135737_010 |
0.1012331697 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |