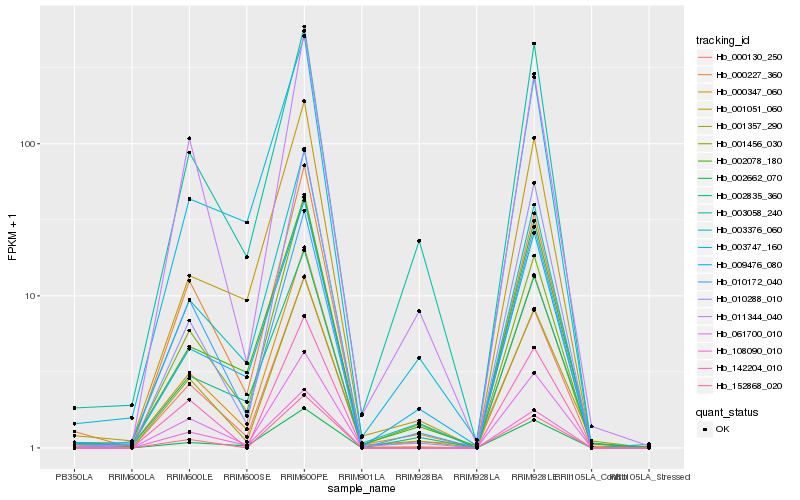
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_250 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |
| 2 |
Hb_061700_010 |
0.0714915018 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 3 |
Hb_000227_360 |
0.0746838738 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 4 |
Hb_011344_040 |
0.0758238713 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 5 |
Hb_001357_290 |
0.0758899246 |
- |
- |
PREDICTED: uncharacterized protein LOC105632986 [Jatropha curcas] |
| 6 |
Hb_000347_060 |
0.0807813618 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 7 |
Hb_003376_060 |
0.0826281385 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 8 |
Hb_108090_010 |
0.0833212731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002662_070 |
0.0849718815 |
- |
- |
hypothetical protein JCGZ_16102 [Jatropha curcas] |
| 10 |
Hb_001456_030 |
0.0887622524 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 11 |
Hb_009476_080 |
0.0906968658 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 12 |
Hb_152868_020 |
0.0951193743 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 13 |
Hb_003058_240 |
0.0951556203 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 14 |
Hb_010288_010 |
0.0954412695 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 15 |
Hb_010172_040 |
0.095688798 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_142204_010 |
0.0962736951 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 17 |
Hb_002835_360 |
0.096843734 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 18 |
Hb_002078_180 |
0.0970160102 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 8 [Jatropha curcas] |
| 19 |
Hb_003747_160 |
0.0990197878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001051_060 |
0.1000983916 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |