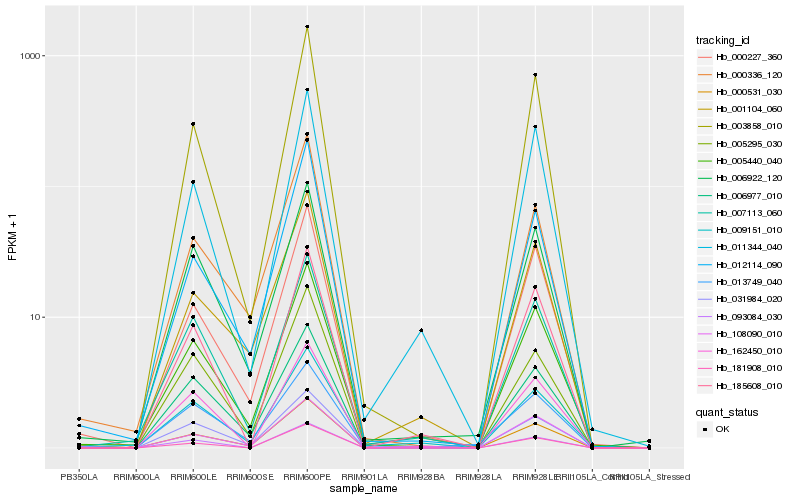
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005440_040 |
0.0 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 2 |
Hb_000227_360 |
0.0617890513 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 3 |
Hb_007113_060 |
0.0680642532 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 4 |
Hb_006922_120 |
0.0689403196 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 5 |
Hb_003858_010 |
0.0709098871 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 6 |
Hb_000531_030 |
0.0744017847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_108090_010 |
0.0760815302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006977_010 |
0.0813433517 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_011344_040 |
0.087193864 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 10 |
Hb_009151_010 |
0.0902498882 |
- |
- |
Peroxidase 3 [Morus notabilis] |
| 11 |
Hb_001104_060 |
0.0918996235 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 12 |
Hb_000336_120 |
0.0982320705 |
- |
- |
PREDICTED: mavicyanin [Populus euphratica] |
| 13 |
Hb_185608_010 |
0.0982658738 |
- |
- |
hypothetical protein POPTR_0004s22210g [Populus trichocarpa] |
| 14 |
Hb_013749_040 |
0.0989110292 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PXL1 [Jatropha curcas] |
| 15 |
Hb_093084_030 |
0.1004621917 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 16 |
Hb_005295_030 |
0.1006369113 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 17 |
Hb_181908_010 |
0.1024760073 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 18 |
Hb_012114_090 |
0.1032986988 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 19 |
Hb_031984_020 |
0.1037228585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_162450_010 |
0.1038141285 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |