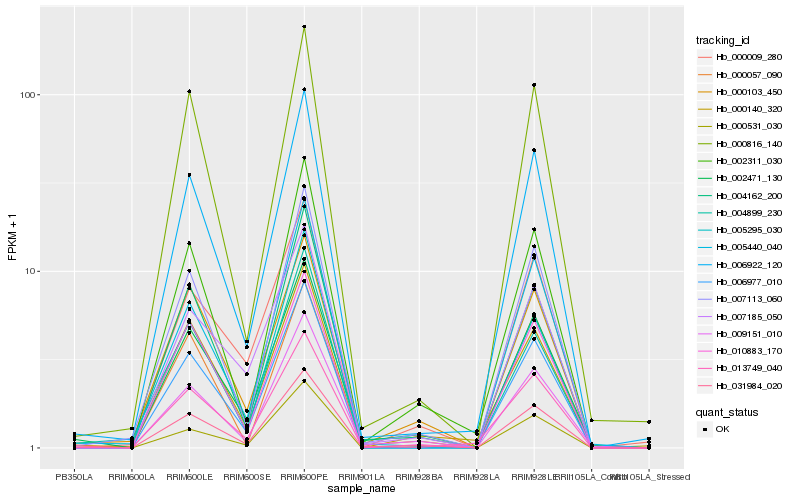
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013749_040 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PXL1 [Jatropha curcas] |
| 2 |
Hb_006922_120 |
0.0606646606 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 3 |
Hb_007113_060 |
0.090941297 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 4 |
Hb_006977_010 |
0.0912404913 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000140_320 |
0.0934608214 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 6 |
Hb_005440_040 |
0.0989110292 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 7 |
Hb_002311_030 |
0.0992106064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000816_140 |
0.1018757091 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 9 |
Hb_010883_170 |
0.1023122813 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 10 |
Hb_002471_130 |
0.1032623102 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 11 |
Hb_031984_020 |
0.1060550318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004162_200 |
0.1070112562 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 13 |
Hb_007185_050 |
0.113300562 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 14 |
Hb_009151_010 |
0.1159730654 |
- |
- |
Peroxidase 3 [Morus notabilis] |
| 15 |
Hb_000009_280 |
0.1173394309 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 16 |
Hb_000531_030 |
0.1182391948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000103_450 |
0.1194513336 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 18 |
Hb_000057_090 |
0.1237483731 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |
| 19 |
Hb_004899_230 |
0.126591655 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 20 |
Hb_005295_030 |
0.1268331159 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |