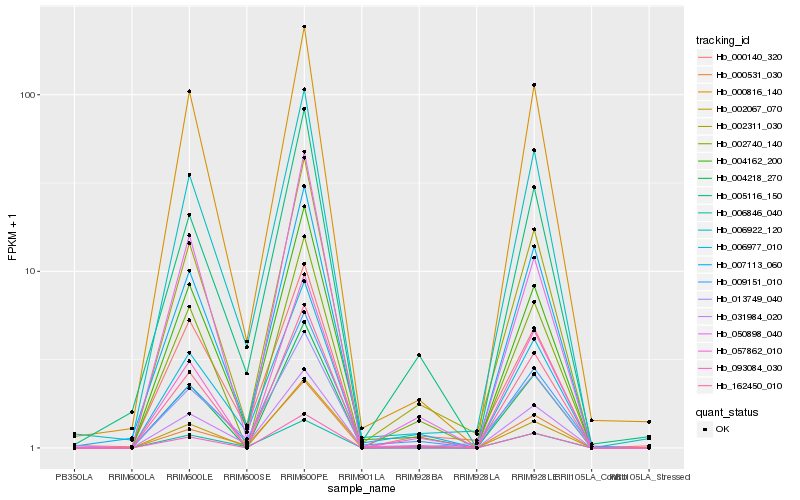
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004162_200 |
0.0638679485 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 3 |
Hb_007113_060 |
0.0696815241 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 4 |
Hb_002740_140 |
0.0711628758 |
- |
- |
hypothetical protein L484_022679 [Morus notabilis] |
| 5 |
Hb_009151_010 |
0.0735825629 |
- |
- |
Peroxidase 3 [Morus notabilis] |
| 6 |
Hb_057862_010 |
0.0795143651 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_162450_010 |
0.082324265 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 8 |
Hb_006922_120 |
0.0845859749 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 9 |
Hb_005116_150 |
0.0891346759 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000140_320 |
0.0901186058 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 11 |
Hb_000816_140 |
0.091108935 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 12 |
Hb_050898_040 |
0.0939611715 |
- |
- |
- |
| 13 |
Hb_002067_070 |
0.0940591462 |
- |
- |
hypothetical protein JCGZ_19254 [Jatropha curcas] |
| 14 |
Hb_031984_020 |
0.0957649424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004218_270 |
0.0965959899 |
- |
- |
hypothetical protein CICLE_v100134152mg, partial [Citrus clementina] |
| 16 |
Hb_000531_030 |
0.0969687741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006846_040 |
0.0976131944 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 18 |
Hb_006977_010 |
0.0983178181 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_093084_030 |
0.0990311891 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 20 |
Hb_013749_040 |
0.0992106064 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PXL1 [Jatropha curcas] |