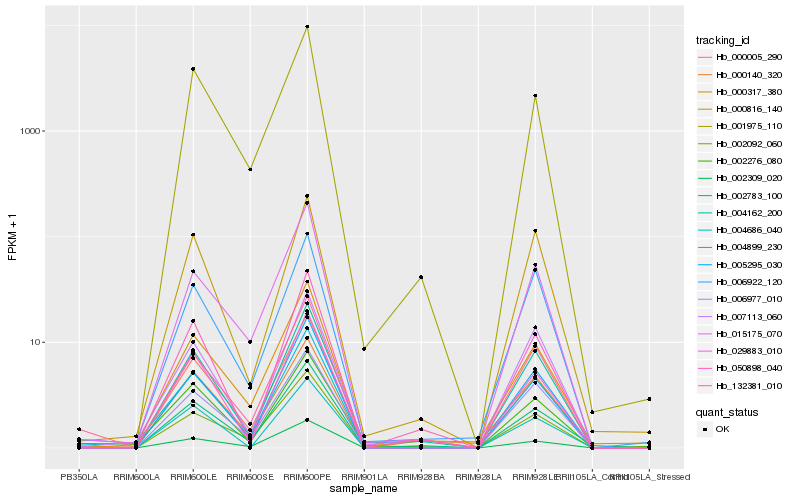
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004899_230 |
0.0 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 2 |
Hb_000005_290 |
0.0732419005 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_005295_030 |
0.0786038063 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 4 |
Hb_002276_080 |
0.084722431 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 5 |
Hb_029883_010 |
0.0914671154 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 6 |
Hb_002092_060 |
0.0930452977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006977_010 |
0.0930719433 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_004162_200 |
0.0940065052 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 9 |
Hb_002309_020 |
0.0957768313 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 10 |
Hb_015175_070 |
0.0991778456 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 11 |
Hb_132381_010 |
0.0994802351 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_000317_380 |
0.1007846303 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 13 |
Hb_007113_060 |
0.1024736919 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 14 |
Hb_006922_120 |
0.1029649951 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 15 |
Hb_002783_100 |
0.1045011793 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 16 |
Hb_001975_110 |
0.1050195724 |
- |
- |
- |
| 17 |
Hb_050898_040 |
0.1082521858 |
- |
- |
- |
| 18 |
Hb_000816_140 |
0.1123556422 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 19 |
Hb_004686_040 |
0.1128327806 |
- |
- |
hypothetical protein POPTR_0016s05690g [Populus trichocarpa] |
| 20 |
Hb_000140_320 |
0.1146540998 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |