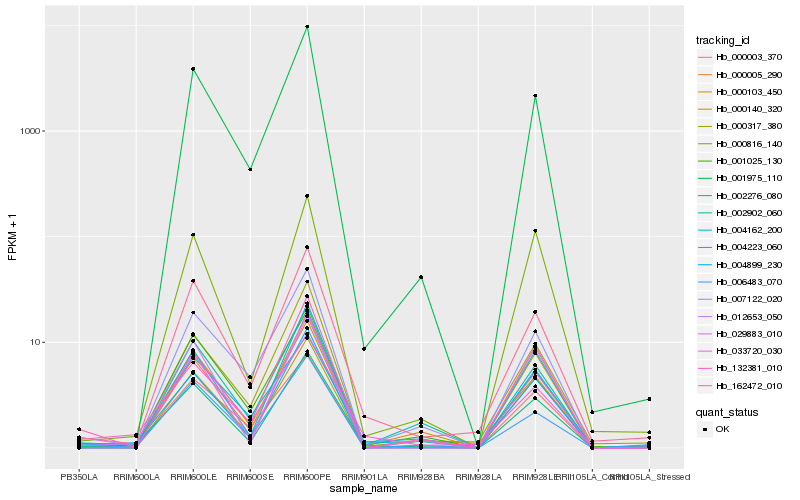
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002276_080 |
0.0 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 2 |
Hb_004899_230 |
0.084722431 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 3 |
Hb_162472_010 |
0.0913553819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006483_070 |
0.1047728196 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_000005_290 |
0.1071602967 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_033720_030 |
0.1085861262 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_004162_200 |
0.1121513954 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 8 |
Hb_001975_110 |
0.1136096924 |
- |
- |
- |
| 9 |
Hb_000140_320 |
0.1146898232 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 10 |
Hb_029883_010 |
0.118564824 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_012653_050 |
0.1206984623 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 12 |
Hb_000317_380 |
0.1210456815 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 13 |
Hb_001025_130 |
0.1220474778 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 14 |
Hb_000103_450 |
0.1231632076 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 15 |
Hb_007122_020 |
0.1233585808 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 16 |
Hb_000816_140 |
0.1243090488 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 17 |
Hb_000003_370 |
0.1246729865 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 18 |
Hb_002902_060 |
0.1247788759 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 19 |
Hb_004223_060 |
0.1255029148 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 20 |
Hb_132381_010 |
0.127695666 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |