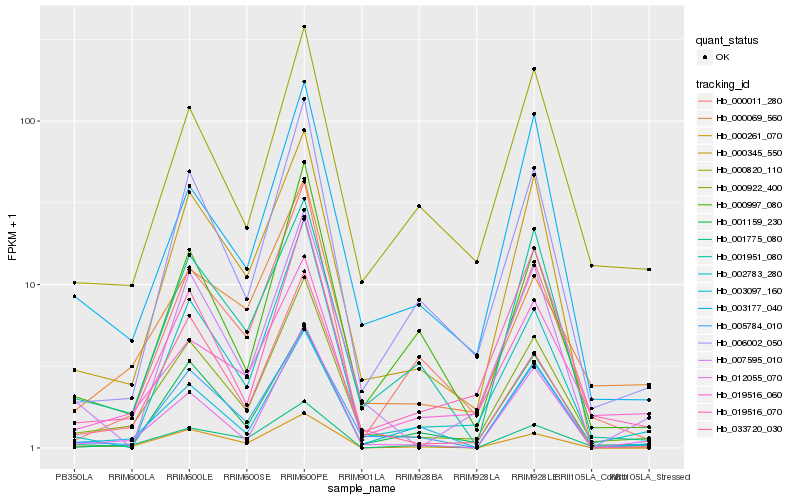
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_400 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 2 |
Hb_006002_050 |
0.1063260597 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 3 |
Hb_000345_550 |
0.1138550497 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 4 |
Hb_033720_030 |
0.1224805781 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_005784_010 |
0.1242111314 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 6 |
Hb_012055_070 |
0.1293392902 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_002783_280 |
0.1302603262 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 8 |
Hb_000820_110 |
0.1307534471 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 9 |
Hb_000997_080 |
0.1323273235 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_019516_070 |
0.1324218227 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 11 |
Hb_000069_560 |
0.1330666529 |
- |
- |
hypothetical protein POPTR_0002s16980g [Populus trichocarpa] |
| 12 |
Hb_007595_010 |
0.134674095 |
- |
- |
Photosystem II reaction center protein K [Medicago truncatula] |
| 13 |
Hb_001159_230 |
0.1354394468 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 14 |
Hb_000011_280 |
0.1360096042 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 15 |
Hb_019516_060 |
0.1385572853 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 16 |
Hb_000261_070 |
0.1397659603 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 17 |
Hb_003097_160 |
0.141002197 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 18 |
Hb_003177_040 |
0.1423406232 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 19 |
Hb_001775_080 |
0.142353161 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 20 |
Hb_001951_080 |
0.1426204119 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |