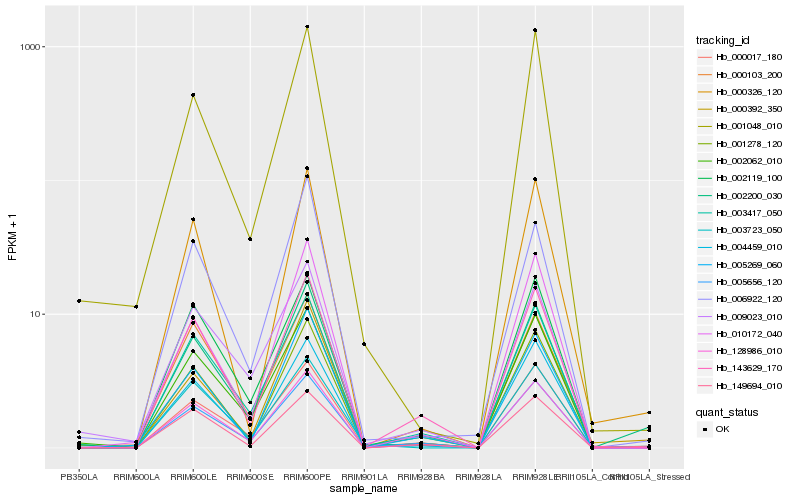
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002200_030 |
0.0 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 2 |
Hb_005656_120 |
0.088613184 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 3 |
Hb_128986_010 |
0.1002825376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003417_050 |
0.1015529956 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002062_010 |
0.105413783 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 6 |
Hb_006922_120 |
0.1074088145 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 7 |
Hb_000392_350 |
0.1080942078 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_001278_120 |
0.1104673525 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 9 |
Hb_004459_010 |
0.1126882859 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 10 |
Hb_000326_120 |
0.116490229 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_009023_010 |
0.1176030861 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 12 |
Hb_000103_200 |
0.1194341105 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 13 |
Hb_001048_010 |
0.1196706555 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |
| 14 |
Hb_149694_010 |
0.120546918 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000017_180 |
0.1209427099 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 16 |
Hb_003723_050 |
0.1209704874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_143629_170 |
0.1216371438 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 18 |
Hb_010172_040 |
0.1220554013 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002119_100 |
0.1223393741 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 20 |
Hb_005269_060 |
0.1228027632 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |