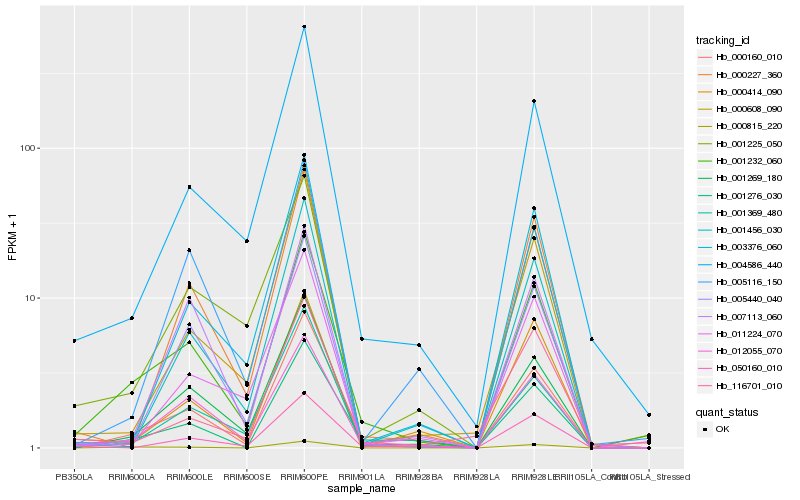
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001232_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012055_070 |
0.1079365126 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_000160_010 |
0.112338417 |
- |
- |
PREDICTED: uncharacterized protein LOC105638816 [Jatropha curcas] |
| 4 |
Hb_001269_180 |
0.1250103889 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_001276_030 |
0.1367904734 |
- |
- |
PREDICTED: uncharacterized protein LOC104881333 [Vitis vinifera] |
| 6 |
Hb_004586_440 |
0.1500344753 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 7 |
Hb_000815_220 |
0.1513534258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001225_050 |
0.1536633683 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 9 |
Hb_001369_480 |
0.1544935128 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0012s14660g [Populus trichocarpa] |
| 10 |
Hb_005440_040 |
0.1608708479 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 11 |
Hb_050160_010 |
0.1612639404 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 12 |
Hb_000227_360 |
0.1646806773 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 13 |
Hb_001456_030 |
0.1683727374 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 14 |
Hb_011224_070 |
0.1695264266 |
- |
- |
PREDICTED: solute carrier family 35 member E3 [Jatropha curcas] |
| 15 |
Hb_005116_150 |
0.1711008452 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000608_090 |
0.1711092858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003376_060 |
0.171779751 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000414_090 |
0.1736003816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007113_060 |
0.175883247 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 20 |
Hb_116701_010 |
0.1760220748 |
- |
- |
hypothetical protein POPTR_0005s19410g [Populus trichocarpa] |