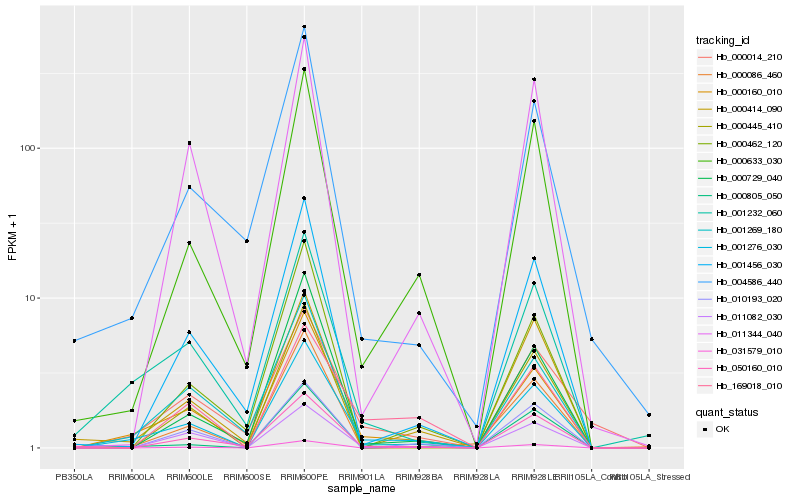
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001276_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104881333 [Vitis vinifera] |
| 2 |
Hb_000160_010 |
0.0583473139 |
- |
- |
PREDICTED: uncharacterized protein LOC105638816 [Jatropha curcas] |
| 3 |
Hb_000633_030 |
0.1312388935 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 4 |
Hb_001232_060 |
0.1367904734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001269_180 |
0.1467961111 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_001456_030 |
0.1598337859 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 7 |
Hb_011082_030 |
0.1614761547 |
- |
- |
hypothetical protein RCOM_0852210 [Ricinus communis] |
| 8 |
Hb_010193_020 |
0.16279708 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 9 |
Hb_050160_010 |
0.1635485099 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 10 |
Hb_000414_090 |
0.1635867139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000805_050 |
0.1639952605 |
- |
- |
zinc transporter, putative [Ricinus communis] |
| 12 |
Hb_169018_010 |
0.1643961229 |
- |
- |
hypothetical protein POPTR_0014s00800g [Populus trichocarpa] |
| 13 |
Hb_000729_040 |
0.1658717702 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |
| 14 |
Hb_031579_010 |
0.1663152933 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 15 |
Hb_011344_040 |
0.1673399392 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 16 |
Hb_000086_460 |
0.1688129019 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 17 |
Hb_004586_440 |
0.1689787352 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 18 |
Hb_000462_120 |
0.1711636964 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 19 |
Hb_000445_410 |
0.1725363084 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |
| 20 |
Hb_000014_210 |
0.1728257219 |
- |
- |
amine oxidase, partial [Aphanizomenon flos-aquae] |