Hb_000633_030
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig633: 56623-59301 |
| Sequence |   |
Annotation
kegg
| ID | pop:POPTR_0003s21020g |
|---|---|
| description | POPTRDRAFT_749226; hypothetical protein |
nr
| ID | XP_002303994.2 |
|---|---|
| description | hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
swissprot
| ID | Q9LVC0 |
|---|---|
| description | Arabinogalactan peptide 14 OS=Arabidopsis thaliana GN=AGP14 PE=1 SV=1 |
trembl
| ID | B9GY63 |
|---|---|
| description | Uncharacterized protein (Fragment) OS=Populus trichocarpa GN=POPTR_0003s21020g PE=4 SV=2 |
Gene Ontology
| ID | GO:0005886 |
|---|---|
| description | arabinogalactan peptide 13-like |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_51865: 58572-59417 |
|---|---|
| cDNA (Sanger) (ID:Location) |
006_M20.ab1: 58685-59419 , 009_I10.ab1: 58781-59417 , 012_C03.ab1: 58786-59417 , 013_I14.ab1: 58855-59417 , 015_P11.ab1: 58773-59420 , 017_L02.ab1: 58754-59419 , 023_J22.ab1: 58756-59419 , 031_K21.ab1: 58789-59416 , 032_J17.ab1: 58914-59415 , 035_C18.ab1: 58688-59419 , 038_L05.ab1: 58632-59415 , 039_B22.ab1: 58954-59417 , 043_C07.ab1: 58827-59413 , 043_M24.ab1: 58693-59415 , 045_A14.ab1: 58673-59415 , 047_E01.ab1: 58734-59413 , 048_K17.ab1: 58687-59415 , 049_C22.ab1: 58896-59394 , 050_C04.ab1: 58785-59415 , 050_G07.ab1: 58797-59413 , 052_F23.ab1: 58713-59417 , 052_P03.ab1: 58771-59415 |
Similar expressed genes (Top20)
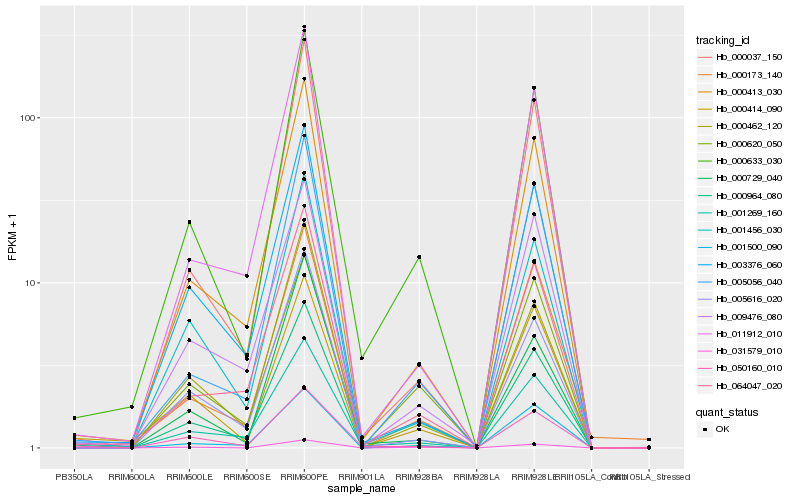
Gene co-expression network
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 0.777415 | 2.45968 | 22.3745 | 336.638 | 0.517896 | 2.48629 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 0 | 0 | 0 | 13.3386 | 151.659 |

