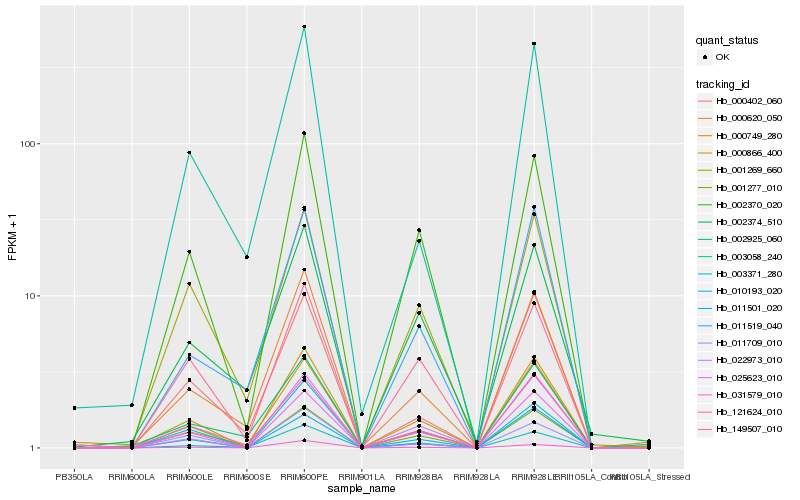
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_400 |
0.0 |
- |
- |
- |
| 2 |
Hb_025623_010 |
0.0546677864 |
- |
- |
- |
| 3 |
Hb_002370_020 |
0.0623203785 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 4 |
Hb_011501_020 |
0.0997047359 |
- |
- |
PREDICTED: uncharacterized protein LOC105648929 [Jatropha curcas] |
| 5 |
Hb_000620_050 |
0.1014283584 |
- |
- |
PREDICTED: uncharacterized protein LOC105649924 [Jatropha curcas] |
| 6 |
Hb_031579_010 |
0.1087609529 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 7 |
Hb_149507_010 |
0.1088493231 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |
| 8 |
Hb_003371_280 |
0.1115065542 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_011709_010 |
0.1123965577 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 10 |
Hb_022973_010 |
0.1148456853 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Jatropha curcas] |
| 11 |
Hb_001269_660 |
0.1209927727 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 12 |
Hb_121624_010 |
0.121627328 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 13 |
Hb_011519_040 |
0.1276599337 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 14 |
Hb_000402_060 |
0.1288142997 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 15 |
Hb_010193_020 |
0.1297401679 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 16 |
Hb_000749_280 |
0.1366340837 |
- |
- |
PREDICTED: uncharacterized protein LOC105138169 [Populus euphratica] |
| 17 |
Hb_002925_060 |
0.1393034216 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 18 |
Hb_002374_510 |
0.1421519983 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 19 |
Hb_003058_240 |
0.1425661972 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 20 |
Hb_001277_010 |
0.1439807658 |
- |
- |
unknown [Populus trichocarpa] |