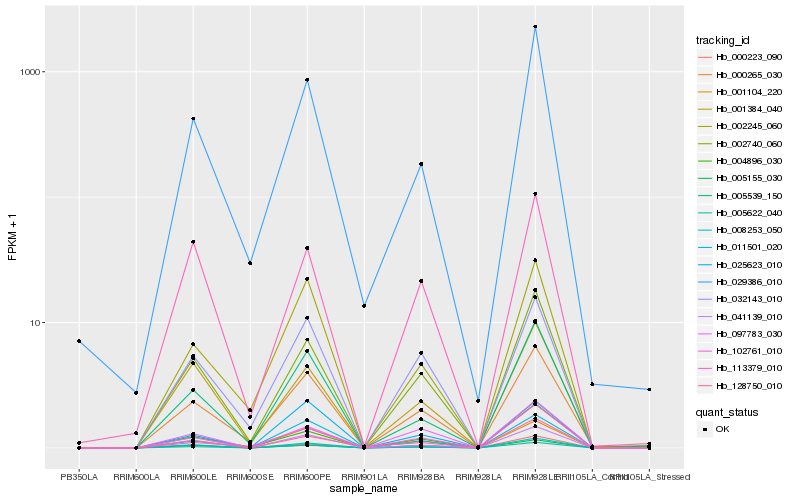
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005622_040 |
0.0 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 2 |
Hb_001104_220 |
0.069178724 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 3 |
Hb_002740_060 |
0.086689482 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 4 |
Hb_000265_030 |
0.0920103044 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 5 |
Hb_008253_050 |
0.0976862165 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 6 |
Hb_097783_030 |
0.0991796757 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 7 |
Hb_011501_020 |
0.1050263412 |
- |
- |
PREDICTED: uncharacterized protein LOC105648929 [Jatropha curcas] |
| 8 |
Hb_032143_010 |
0.1061841315 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 9 |
Hb_005155_030 |
0.1158995814 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 10 |
Hb_004896_030 |
0.1192963433 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 11 |
Hb_005539_150 |
0.1194732248 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 12 |
Hb_128750_010 |
0.1275962666 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_002245_060 |
0.1326254878 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_041139_010 |
0.133352242 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 15 |
Hb_000223_090 |
0.1350351558 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 16 |
Hb_029386_010 |
0.1357442516 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 17 |
Hb_113379_010 |
0.1371807358 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 18 |
Hb_001384_040 |
0.1389948925 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 19 |
Hb_025623_010 |
0.1397211428 |
- |
- |
- |
| 20 |
Hb_102761_010 |
0.1410144648 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |