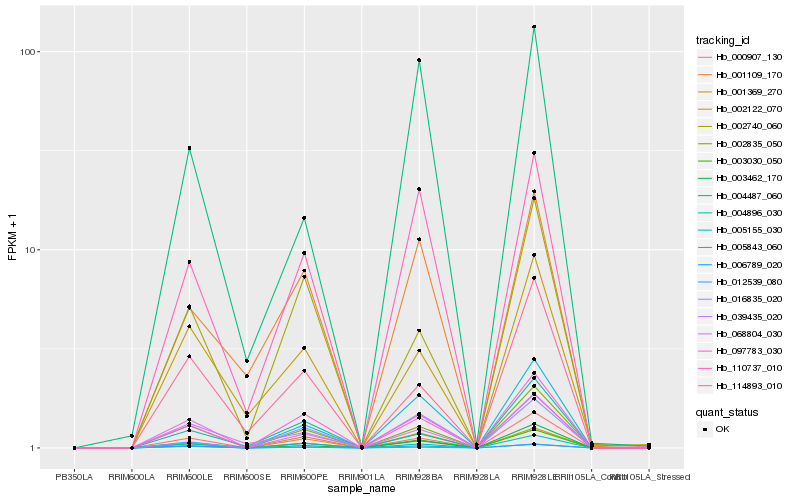
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003030_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_097783_030 |
0.0818871991 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 3 |
Hb_002835_050 |
0.082045533 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 4 |
Hb_000907_130 |
0.0876226075 |
- |
- |
- |
| 5 |
Hb_005155_030 |
0.0915494613 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_110737_010 |
0.1002759366 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 7 |
Hb_001369_270 |
0.1372143397 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_068804_030 |
0.137738758 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 9 |
Hb_001109_170 |
0.1469378239 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_016835_020 |
0.1494858985 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_004487_060 |
0.1613746746 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_039435_020 |
0.1626821133 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 13 |
Hb_012539_080 |
0.16440711 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 14 |
Hb_006789_020 |
0.1648876495 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase-like 22 [Camelina sativa] |
| 15 |
Hb_002740_060 |
0.1673128161 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 16 |
Hb_003462_170 |
0.1713949644 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_114893_010 |
0.1718002035 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_004896_030 |
0.1732691101 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 19 |
Hb_005843_060 |
0.1733676647 |
- |
- |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X3 [Nelumbo nucifera] |
| 20 |
Hb_002122_070 |
0.173655823 |
- |
- |
unnamed protein product [Vitis vinifera] |