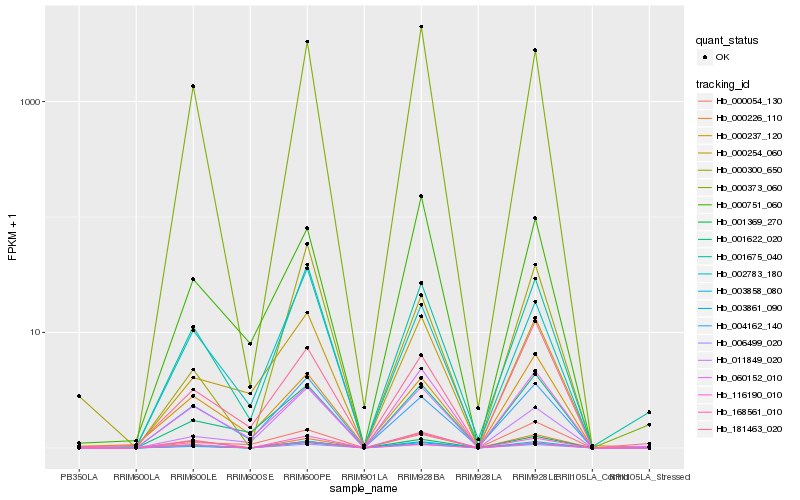
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003858_080 |
0.0 |
- |
- |
Germin-like protein 2 [Theobroma cacao] |
| 2 |
Hb_003861_090 |
0.1040931028 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 3 |
Hb_001675_040 |
0.114926463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001369_270 |
0.122495841 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_006499_020 |
0.1271436801 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 6 |
Hb_000373_060 |
0.1313762754 |
- |
- |
PREDICTED: extensin-3 [Camelina sativa] |
| 7 |
Hb_168561_010 |
0.1380080037 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |
| 8 |
Hb_001622_020 |
0.1507352988 |
- |
- |
- |
| 9 |
Hb_116190_010 |
0.1528250326 |
- |
- |
PREDICTED: uncharacterized protein LOC105118535 [Populus euphratica] |
| 10 |
Hb_000254_060 |
0.157255209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_060152_010 |
0.1611554944 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 12 |
Hb_004162_140 |
0.1613263364 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 13 |
Hb_000226_110 |
0.1625004189 |
- |
- |
- |
| 14 |
Hb_002783_180 |
0.1649102972 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 15 |
Hb_181463_020 |
0.1715672051 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 16 |
Hb_000054_130 |
0.1740916692 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 17 |
Hb_000300_650 |
0.1744632484 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 18 |
Hb_000237_120 |
0.1780405237 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 19 |
Hb_011849_020 |
0.1812275886 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 20 |
Hb_000751_060 |
0.1823746358 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |