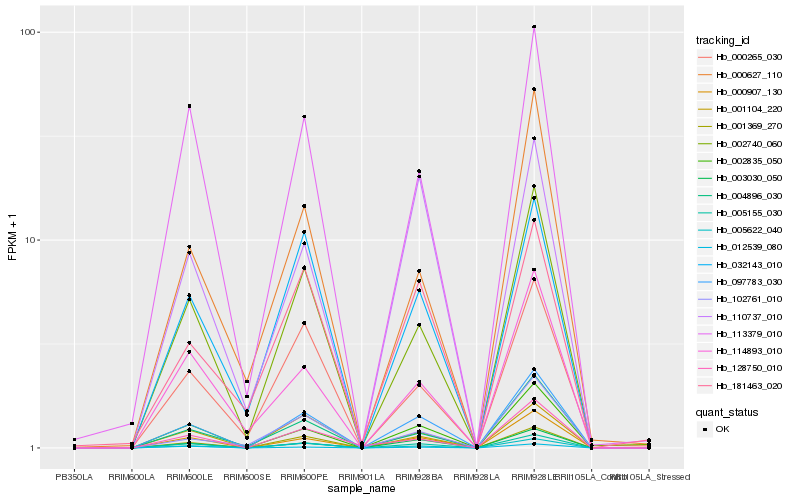
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_097783_030 |
0.0 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 2 |
Hb_005155_030 |
0.0534728349 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 3 |
Hb_000907_130 |
0.0778684736 |
- |
- |
- |
| 4 |
Hb_003030_050 |
0.0818871991 |
- |
- |
- |
| 5 |
Hb_002835_050 |
0.0824939858 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 6 |
Hb_005622_040 |
0.0991796757 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 7 |
Hb_002740_060 |
0.1034830167 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 8 |
Hb_004896_030 |
0.1204135809 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 9 |
Hb_113379_010 |
0.1374808347 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 10 |
Hb_032143_010 |
0.1390229513 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_128750_010 |
0.1420584826 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 12 |
Hb_000265_030 |
0.1464206203 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 13 |
Hb_001369_270 |
0.1464260664 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001104_220 |
0.1468464873 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 15 |
Hb_114893_010 |
0.1492156349 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 16 |
Hb_110737_010 |
0.1527362669 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 17 |
Hb_181463_020 |
0.1533365828 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 18 |
Hb_012539_080 |
0.1534727777 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 19 |
Hb_102761_010 |
0.1542257955 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 20 |
Hb_000627_110 |
0.1544418191 |
- |
- |
cellulose synthase, putative [Ricinus communis] |