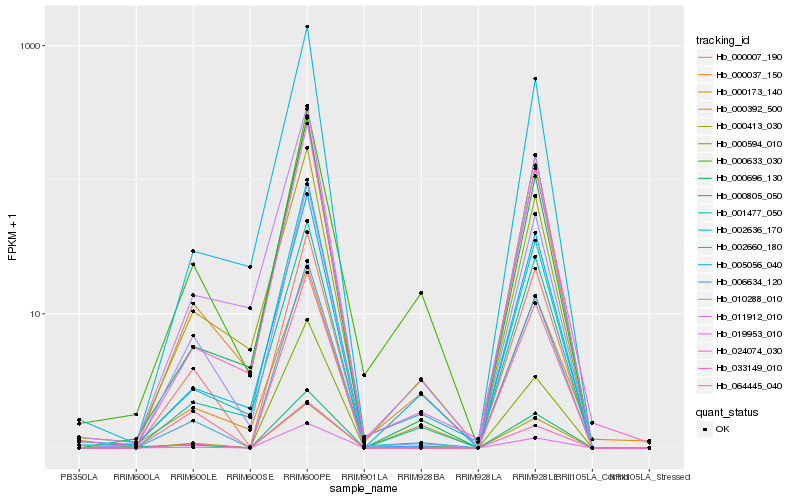
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_050 |
0.0 |
- |
- |
zinc transporter, putative [Ricinus communis] |
| 2 |
Hb_006634_120 |
0.0846646866 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 3 |
Hb_000037_150 |
0.0923115344 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 4 |
Hb_005056_040 |
0.0940726974 |
- |
- |
hypothetical protein POPTR_0007s14730g [Populus trichocarpa] |
| 5 |
Hb_064445_040 |
0.0953075493 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |
| 6 |
Hb_001477_050 |
0.0970188161 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_024074_030 |
0.1007854913 |
- |
- |
hypothetical protein CISIN_1g043166mg [Citrus sinensis] |
| 8 |
Hb_019953_010 |
0.1022275915 |
- |
- |
hypothetical protein CICLE_v10019877mg [Citrus clementina] |
| 9 |
Hb_033149_010 |
0.1056352958 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_000696_130 |
0.106672178 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_000007_190 |
0.1088075903 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 10-like [Jatropha curcas] |
| 12 |
Hb_010288_010 |
0.1101459736 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 13 |
Hb_002636_170 |
0.1109436731 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 14 |
Hb_000392_500 |
0.1112722878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002660_180 |
0.1142900215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000173_140 |
0.1184837502 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 17 |
Hb_011912_010 |
0.1195662465 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000413_030 |
0.1206689375 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 19 |
Hb_000594_010 |
0.1218008544 |
- |
- |
PREDICTED: lysine histidine transporter-like 6 isoform X3 [Populus euphratica] |
| 20 |
Hb_000633_030 |
0.1229122117 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |