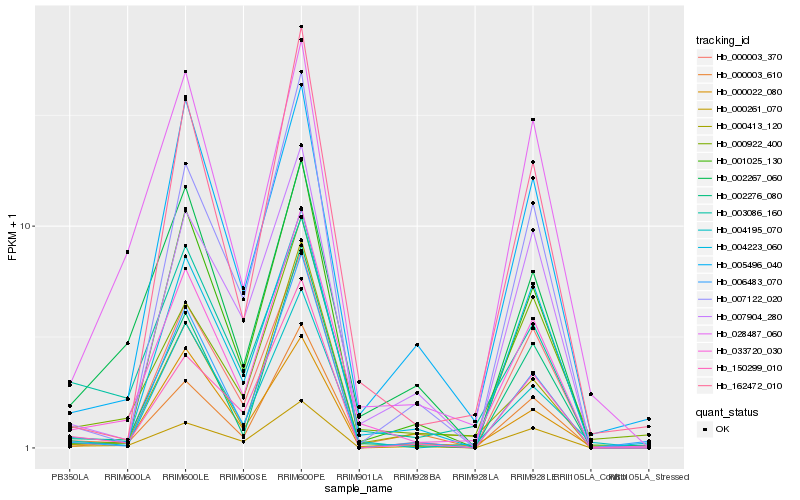
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033720_030 |
0.0 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_002276_080 |
0.1085861262 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 3 |
Hb_162472_010 |
0.1094359906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004195_070 |
0.1142309855 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_004223_060 |
0.1191338366 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 6 |
Hb_000922_400 |
0.1224805781 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 7 |
Hb_001025_130 |
0.1276799174 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 8 |
Hb_000261_070 |
0.1281504019 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 9 |
Hb_150299_010 |
0.1310349 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Tarenaya hassleriana] |
| 10 |
Hb_000003_610 |
0.1324394028 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase [Jatropha curcas] |
| 11 |
Hb_000003_370 |
0.1327396734 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 12 |
Hb_028487_060 |
0.1355276591 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 13 |
Hb_003086_160 |
0.1362821994 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |
| 14 |
Hb_000022_080 |
0.1363067479 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 15 |
Hb_007904_280 |
0.1385143957 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 16 |
Hb_000413_120 |
0.1391334229 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 17 |
Hb_007122_020 |
0.1395260841 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 18 |
Hb_006483_070 |
0.139611792 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_002267_060 |
0.1412024723 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 20 |
Hb_005496_040 |
0.14496041 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |