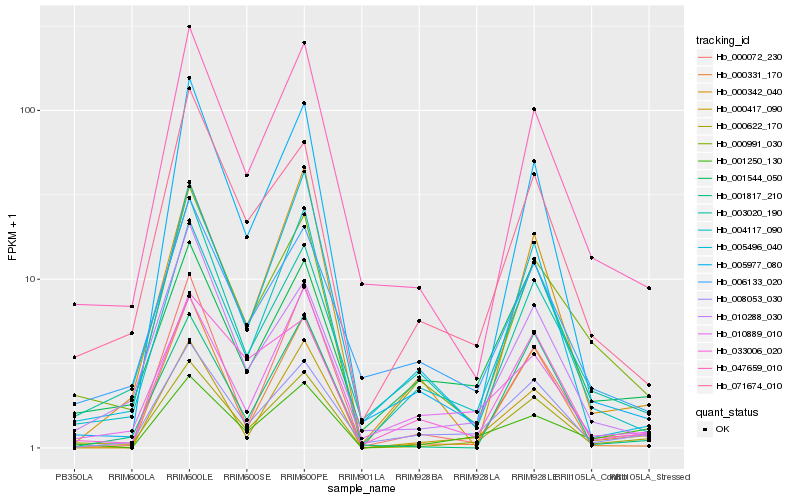
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000622_170 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 2 |
Hb_047659_010 |
0.1251834948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000991_030 |
0.1263954654 |
- |
- |
PREDICTED: uncharacterized protein LOC105122436 [Populus euphratica] |
| 4 |
Hb_008053_030 |
0.1284154442 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 5 |
Hb_004117_090 |
0.1335028746 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 6 |
Hb_000417_090 |
0.1445634733 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 7 |
Hb_071674_010 |
0.1446431549 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 8 |
Hb_001250_130 |
0.1528763354 |
- |
- |
hypothetical protein POPTR_0008s19280g [Populus trichocarpa] |
| 9 |
Hb_000072_230 |
0.1538086693 |
- |
- |
hypothetical protein POPTR_0004s10790g [Populus trichocarpa] |
| 10 |
Hb_010889_010 |
0.1567667089 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 11 |
Hb_010288_030 |
0.1578116561 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 12 |
Hb_001817_210 |
0.1601868406 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005496_040 |
0.1606320435 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 14 |
Hb_000342_040 |
0.16092694 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 15 |
Hb_005977_080 |
0.1615229212 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 16 |
Hb_001544_050 |
0.1622930444 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 17 |
Hb_003020_190 |
0.162833215 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 18 |
Hb_000331_170 |
0.1628397208 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 19 |
Hb_006133_020 |
0.1639010452 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 20 |
Hb_033006_020 |
0.1649402969 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |