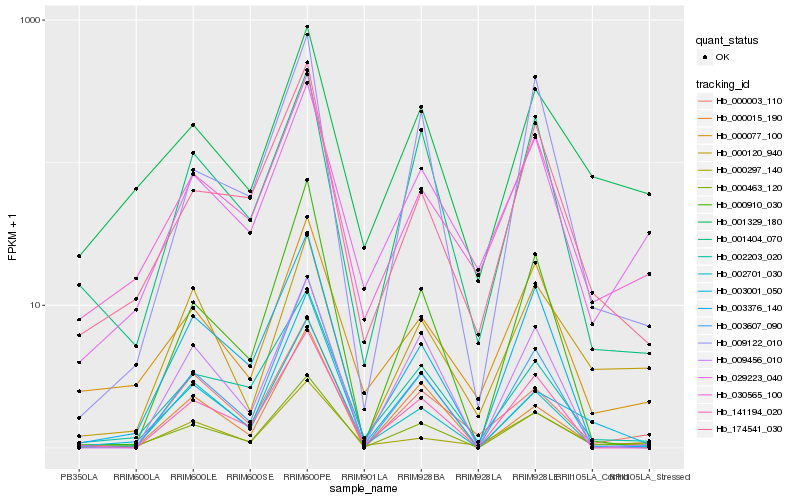
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000463_120 |
0.0 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 2 |
Hb_030565_100 |
0.1322399517 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 3 |
Hb_001404_070 |
0.1362237331 |
- |
- |
sucrose synthase 4 [Hevea brasiliensis] |
| 4 |
Hb_002203_020 |
0.1388156675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_029223_040 |
0.1422202378 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 6 |
Hb_000077_100 |
0.1437250702 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_009122_010 |
0.145581751 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_174541_030 |
0.1457444982 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 9 |
Hb_001329_180 |
0.1461736742 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 10 |
Hb_002701_030 |
0.1479964125 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 11 |
Hb_003376_140 |
0.149662868 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 12 |
Hb_000015_190 |
0.1498811698 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 13 |
Hb_000297_140 |
0.1502256782 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 14 |
Hb_141194_020 |
0.1503052279 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 15 |
Hb_003607_090 |
0.1510700059 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 16 |
Hb_000120_940 |
0.1521704633 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000003_110 |
0.1534571897 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000910_030 |
0.1548458109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009456_010 |
0.1589918748 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |
| 20 |
Hb_003001_050 |
0.1593741949 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |