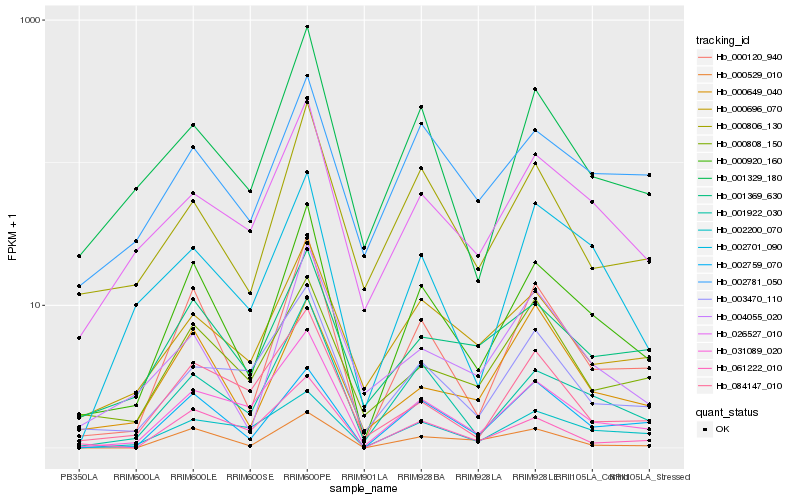
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_160 |
0.0 |
- |
- |
annexin [Manihot esculenta] |
| 2 |
Hb_000120_940 |
0.0869817859 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_001922_030 |
0.1228139003 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 4 |
Hb_031089_020 |
0.1380835923 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_002759_070 |
0.1435952672 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 6 |
Hb_001329_180 |
0.1540834879 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 7 |
Hb_026527_010 |
0.1568114991 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 8 |
Hb_001369_630 |
0.1625813662 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 9 |
Hb_000529_010 |
0.1629919767 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 10 |
Hb_000806_130 |
0.1674504594 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 11 |
Hb_061222_010 |
0.1696517203 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 12 |
Hb_000696_070 |
0.171989289 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 13 |
Hb_002701_090 |
0.1733175998 |
- |
- |
hypothetical protein POPTR_0017s13430g [Populus trichocarpa] |
| 14 |
Hb_004055_020 |
0.1744092101 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 15 |
Hb_002200_070 |
0.1750649733 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 16 |
Hb_002781_050 |
0.176950378 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 17 |
Hb_003470_110 |
0.177310776 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000808_150 |
0.178870631 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 19 |
Hb_084147_010 |
0.1792513759 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_000649_040 |
0.1805200288 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |