| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
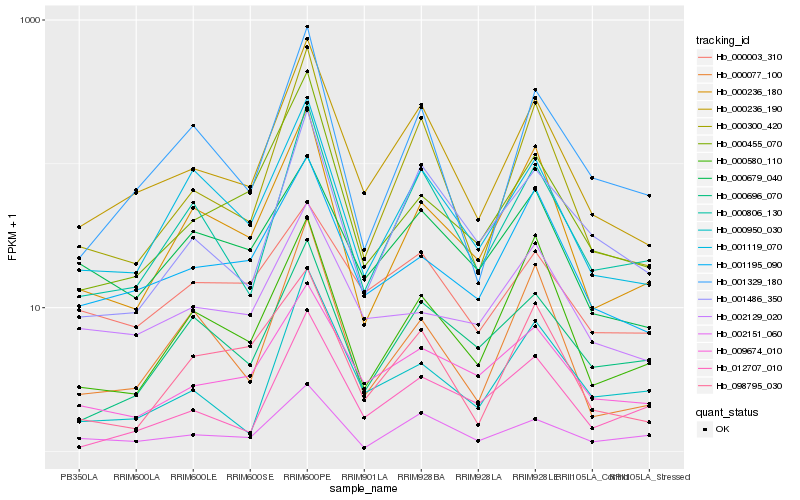
Hb_000236_190 |
0.0 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 2 |
Hb_000300_420 |
0.1048716538 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000806_130 |
0.1086939576 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 4 |
Hb_001119_070 |
0.1271724276 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 5 |
Hb_001486_350 |
0.129631333 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001329_180 |
0.1310697241 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 7 |
Hb_009674_010 |
0.1365162415 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 8 |
Hb_001195_090 |
0.1384154909 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 9 |
Hb_098795_030 |
0.1478870141 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 10 |
Hb_000236_180 |
0.1487612443 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 11 |
Hb_002151_060 |
0.1500154836 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc4 [Jatropha curcas] |
| 12 |
Hb_000679_040 |
0.1528301731 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 13 |
Hb_000696_070 |
0.1543159651 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 14 |
Hb_000455_070 |
0.1546276968 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 15 |
Hb_000077_100 |
0.1579217575 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_012707_010 |
0.1596771838 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 17 |
Hb_000950_030 |
0.159863938 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 18 |
Hb_000580_110 |
0.160656574 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 19 |
Hb_002129_020 |
0.1632819885 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 20 |
Hb_000003_310 |
0.1662631894 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |