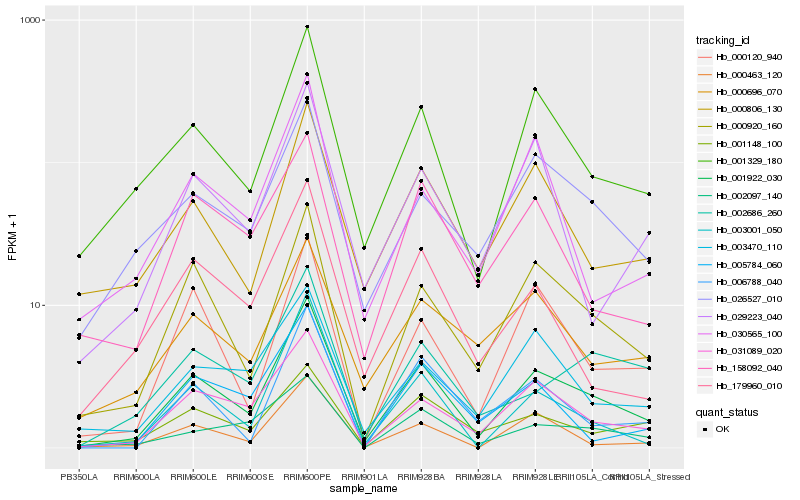
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001922_030 |
0.0 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 2 |
Hb_031089_020 |
0.1199985867 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 3 |
Hb_000920_160 |
0.1228139003 |
- |
- |
annexin [Manihot esculenta] |
| 4 |
Hb_001329_180 |
0.127010279 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 5 |
Hb_000120_940 |
0.1558358982 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_026527_010 |
0.1581691743 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 7 |
Hb_000463_120 |
0.1607187553 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 8 |
Hb_003001_050 |
0.1619687081 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 9 |
Hb_000806_130 |
0.1638152439 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 10 |
Hb_003470_110 |
0.1644818248 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_002097_140 |
0.1684034381 |
- |
- |
CHY zinc finger family protein, expressed [Oryza sativa Japonica Group] |
| 12 |
Hb_006788_040 |
0.1696495837 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 13 |
Hb_005784_060 |
0.1722693444 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 14 |
Hb_002686_260 |
0.1725540333 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 15 |
Hb_029223_040 |
0.1730705032 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 16 |
Hb_158092_040 |
0.1744306402 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 17 |
Hb_179960_010 |
0.1750424554 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 18 |
Hb_001148_100 |
0.1763761232 |
- |
- |
- |
| 19 |
Hb_030565_100 |
0.1789240007 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000696_070 |
0.1792071825 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |