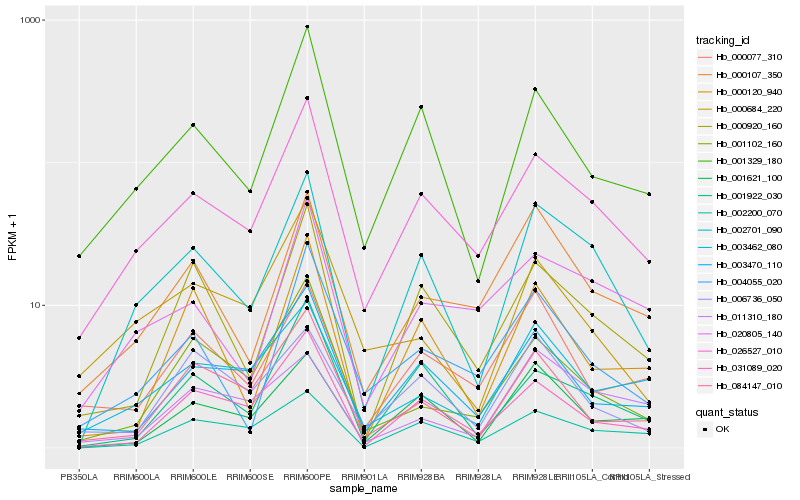
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s13430g [Populus trichocarpa] |
| 2 |
Hb_026527_010 |
0.137245869 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 3 |
Hb_001329_180 |
0.1679645831 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 4 |
Hb_000920_160 |
0.1733175998 |
- |
- |
annexin [Manihot esculenta] |
| 5 |
Hb_002200_070 |
0.175901792 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 6 |
Hb_001621_100 |
0.1765786047 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4 [Jatropha curcas] |
| 7 |
Hb_000107_350 |
0.1813234564 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 8 |
Hb_001922_030 |
0.1872161238 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 9 |
Hb_031089_020 |
0.1916359526 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 10 |
Hb_003470_110 |
0.2026827081 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_006736_050 |
0.2036389307 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 12 |
Hb_004055_020 |
0.2042347447 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 13 |
Hb_020805_140 |
0.2045619907 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 14 |
Hb_003462_080 |
0.2060895643 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000120_940 |
0.2077350586 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_084147_010 |
0.2094260525 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_011310_180 |
0.2118526651 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001102_160 |
0.2134494636 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 19 |
Hb_000684_220 |
0.2155690243 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000077_310 |
0.2157982679 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |