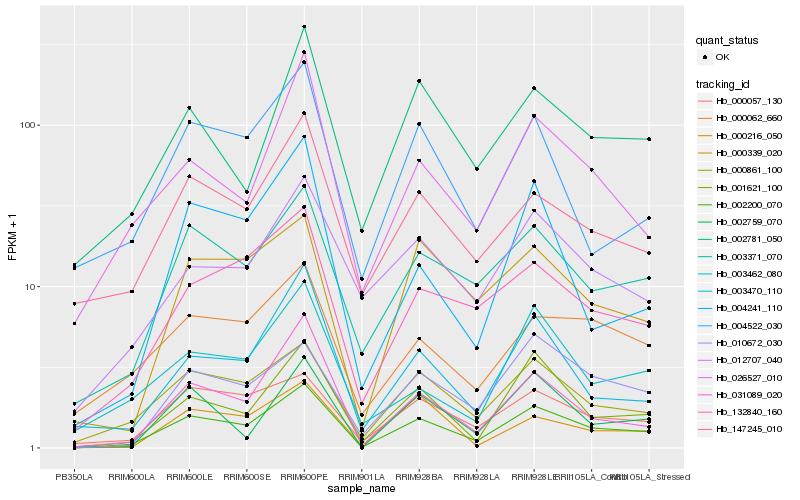
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002200_070 |
0.0 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 2 |
Hb_001621_100 |
0.1255784132 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4 [Jatropha curcas] |
| 3 |
Hb_000861_100 |
0.1285042087 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_031089_020 |
0.1298023604 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_003371_070 |
0.1415087384 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000057_130 |
0.1473456692 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 7 |
Hb_003470_110 |
0.1497728895 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_132840_160 |
0.1513489375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010672_030 |
0.1529809036 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 10 |
Hb_000339_020 |
0.1576521981 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 11 |
Hb_026527_010 |
0.1607383779 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 12 |
Hb_002781_050 |
0.1668266038 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 13 |
Hb_002759_070 |
0.1674797038 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 14 |
Hb_147245_010 |
0.1674823338 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 15 |
Hb_000216_050 |
0.1689966962 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 16 |
Hb_003462_080 |
0.1693498949 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_004241_110 |
0.170438711 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 18 |
Hb_000062_660 |
0.1706589573 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 19 |
Hb_004522_030 |
0.1716298864 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 20 |
Hb_012707_040 |
0.1719784835 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 29 [Jatropha curcas] |