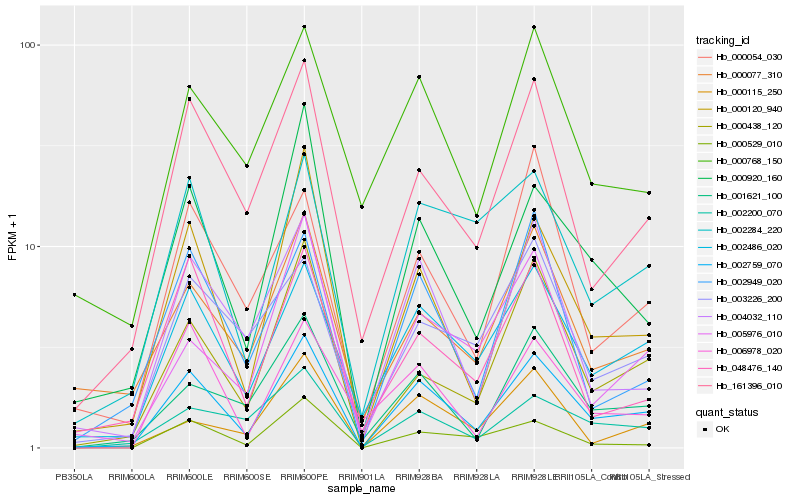
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002759_070 |
0.0 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 2 |
Hb_002486_020 |
0.1276002572 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000120_940 |
0.1325959257 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000920_160 |
0.1435952672 |
- |
- |
annexin [Manihot esculenta] |
| 5 |
Hb_161396_010 |
0.1488780775 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 6 |
Hb_000438_120 |
0.1526572094 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 7 |
Hb_001621_100 |
0.1555069153 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4 [Jatropha curcas] |
| 8 |
Hb_004032_110 |
0.1556131877 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002284_220 |
0.163640995 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 10 |
Hb_002200_070 |
0.1674797038 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 11 |
Hb_000529_010 |
0.167879407 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 12 |
Hb_002949_020 |
0.1680384212 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 13 |
Hb_000077_310 |
0.1728467611 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 14 |
Hb_000054_030 |
0.1754409324 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 15 |
Hb_006978_020 |
0.1757945771 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_000768_150 |
0.1774578135 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 17 |
Hb_005976_010 |
0.1778690503 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_048476_140 |
0.1779267544 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 19 |
Hb_000115_250 |
0.1786857644 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 20 |
Hb_003226_200 |
0.1797085619 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |