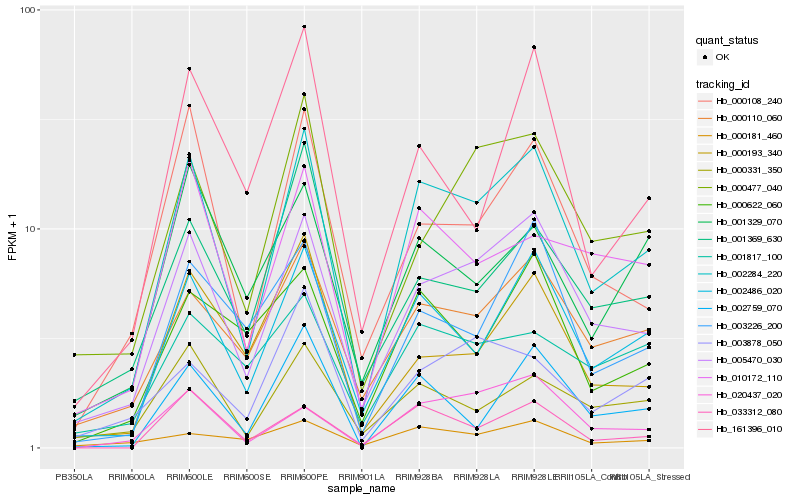
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_220 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 2 |
Hb_005470_030 |
0.0959014346 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002486_020 |
0.1089736024 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000110_060 |
0.1456767629 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 5 |
Hb_033312_080 |
0.1565796945 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 6 |
Hb_000108_240 |
0.1585705529 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 7 |
Hb_010172_110 |
0.1587260205 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 8 |
Hb_000331_350 |
0.1600527453 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 9 |
Hb_000477_040 |
0.1601273459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002759_070 |
0.163640995 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 11 |
Hb_020437_020 |
0.1693876332 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 12 |
Hb_003226_200 |
0.1700574815 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 13 |
Hb_000622_060 |
0.1786482102 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001369_630 |
0.1811682216 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 15 |
Hb_001329_070 |
0.1826859039 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 16 |
Hb_000193_340 |
0.1836353249 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001817_100 |
0.1838521498 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 18 |
Hb_000181_460 |
0.184755057 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 19 |
Hb_003878_050 |
0.1860513263 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 20 |
Hb_161396_010 |
0.1873863798 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |