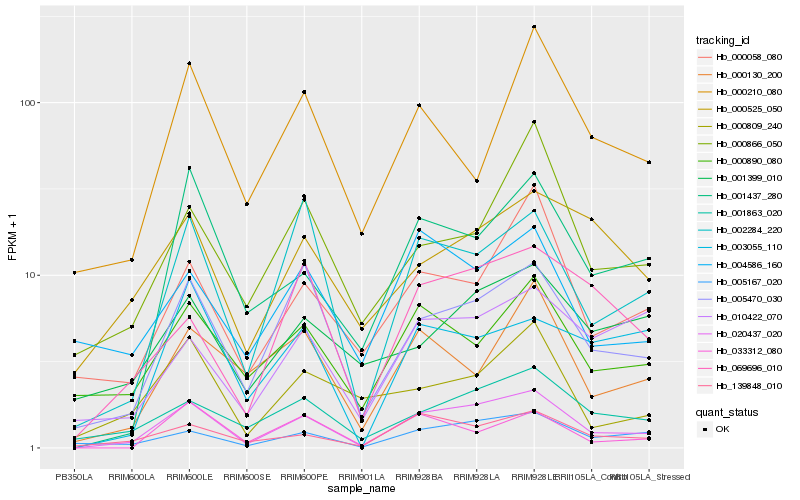
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020437_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 2 |
Hb_005470_030 |
0.1494135003 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002284_220 |
0.1693876332 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 4 |
Hb_005167_020 |
0.1715504836 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 5 |
Hb_139848_010 |
0.1722617637 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |
| 6 |
Hb_001863_020 |
0.1812915104 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001437_280 |
0.1851748039 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 8 |
Hb_069696_010 |
0.1927990889 |
- |
- |
hypothetical protein POPTR_0003s21190g [Populus trichocarpa] |
| 9 |
Hb_000890_080 |
0.1992870651 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 10 |
Hb_000058_080 |
0.2038384656 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_000130_200 |
0.207122146 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 12 |
Hb_033312_080 |
0.2093833366 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 13 |
Hb_000809_240 |
0.2149954703 |
- |
- |
hypothetical protein POPTR_0007s11280g [Populus trichocarpa] |
| 14 |
Hb_003055_110 |
0.2153319586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000525_050 |
0.2168429382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000866_050 |
0.2174040574 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 17 |
Hb_010422_070 |
0.2176484519 |
- |
- |
PREDICTED: UPF0483 protein CBG03338-like [Jatropha curcas] |
| 18 |
Hb_004586_160 |
0.2182310247 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001399_010 |
0.2225430338 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000210_080 |
0.2227809379 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |