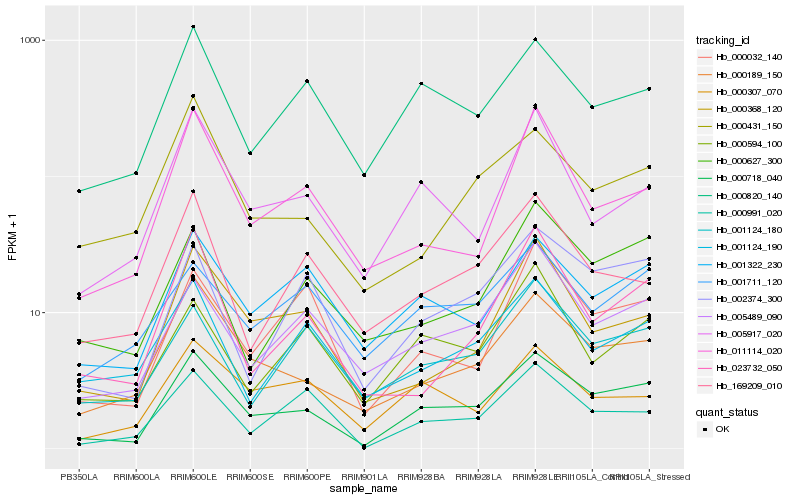
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000718_040 |
0.0 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 2 |
Hb_000189_150 |
0.1320907307 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 3 |
Hb_002374_300 |
0.1498879111 |
- |
- |
hypothetical protein JCGZ_12140 [Jatropha curcas] |
| 4 |
Hb_000820_140 |
0.1524025475 |
- |
- |
histone H4 [Zea mays] |
| 5 |
Hb_001322_230 |
0.1548116869 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 6 |
Hb_005489_090 |
0.1600062545 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_005917_020 |
0.1628855238 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 8 |
Hb_000991_020 |
0.1679456689 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_000368_120 |
0.1680783565 |
- |
- |
PREDICTED: uncharacterized protein LOC105633862 [Jatropha curcas] |
| 10 |
Hb_000627_300 |
0.1712993818 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 11 |
Hb_000431_150 |
0.1716686229 |
- |
- |
ribosomal protein L27, putative [Ricinus communis] |
| 12 |
Hb_000307_070 |
0.1735187766 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 13 |
Hb_001124_180 |
0.173786638 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 14 |
Hb_000032_140 |
0.1748710194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_023732_050 |
0.1766527643 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 16 |
Hb_011114_020 |
0.1767307802 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000594_100 |
0.1768916536 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 18 |
Hb_169209_010 |
0.1786154584 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001124_190 |
0.1792967555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001711_120 |
0.1795213447 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |