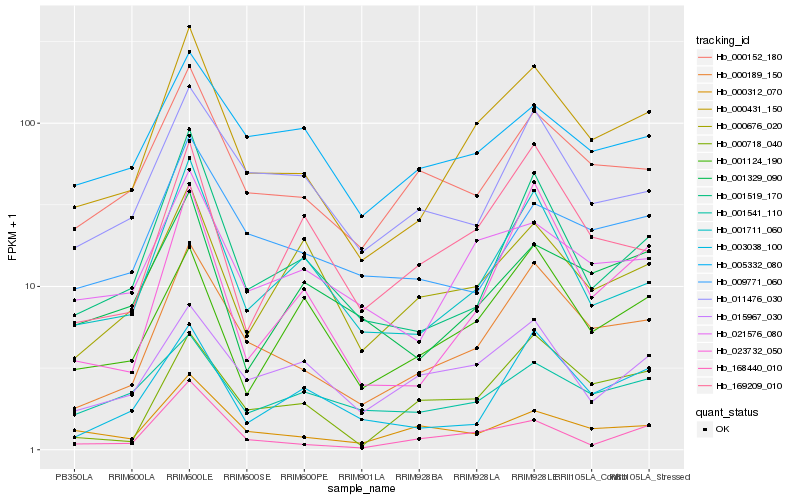
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_150 |
0.0 |
- |
- |
ribosomal protein L27, putative [Ricinus communis] |
| 2 |
Hb_000189_150 |
0.0962889104 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 3 |
Hb_021576_080 |
0.1302983863 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 4 |
Hb_001711_060 |
0.140008663 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 5 |
Hb_001519_170 |
0.1422416257 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000152_180 |
0.1465485087 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 7 |
Hb_023732_050 |
0.1642919097 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 8 |
Hb_000312_070 |
0.1666552889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_168440_010 |
0.1675094001 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 10 |
Hb_001329_090 |
0.1678952004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001124_190 |
0.1679324563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011476_030 |
0.1709935896 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 13 |
Hb_015967_030 |
0.1714463133 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000718_040 |
0.1716686229 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 15 |
Hb_000676_020 |
0.1739252138 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 16 |
Hb_169209_010 |
0.1745533162 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001541_110 |
0.1766967378 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_003038_100 |
0.1770141762 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_009771_060 |
0.1770583252 |
- |
- |
PREDICTED: 30S ribosomal protein S31, chloroplastic [Populus euphratica] |
| 20 |
Hb_005332_080 |
0.1776203723 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |