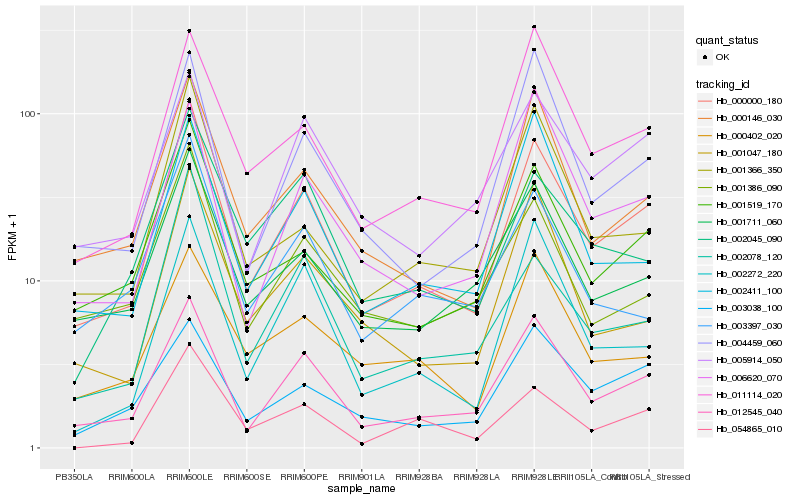
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_180 |
0.0 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 2 |
Hb_012545_040 |
0.0768630336 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_001519_170 |
0.11143829 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000146_030 |
0.1118234924 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 5 |
Hb_011114_020 |
0.1228342351 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_001711_060 |
0.1274831128 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 7 |
Hb_004459_060 |
0.1287118882 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001366_350 |
0.1346486453 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 9 |
Hb_003038_100 |
0.1358288685 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001386_090 |
0.1389772526 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 11 |
Hb_002078_120 |
0.1400859217 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 12 |
Hb_002411_100 |
0.1405640215 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 13 |
Hb_001047_180 |
0.1410856899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005914_050 |
0.142223836 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 15 |
Hb_054865_010 |
0.1432196527 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 16 |
Hb_002045_090 |
0.145371582 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 17 |
Hb_002272_220 |
0.1465591983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006620_070 |
0.1473337674 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 19 |
Hb_003397_030 |
0.1478813729 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000402_020 |
0.1490841831 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |