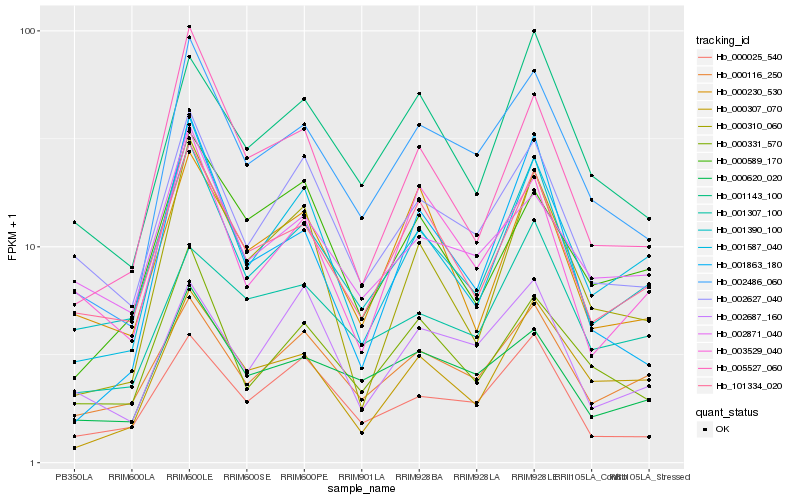
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002486_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 2 |
Hb_000620_020 |
0.1099074218 |
- |
- |
PREDICTED: uncharacterized protein LOC105649917 [Jatropha curcas] |
| 3 |
Hb_000331_570 |
0.1178561647 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 4 |
Hb_001390_100 |
0.1214466831 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002627_040 |
0.1227612265 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 6 |
Hb_101334_020 |
0.1256557457 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 7 |
Hb_001143_100 |
0.1294532599 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 8 |
Hb_005527_060 |
0.1320922998 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 9 |
Hb_000025_540 |
0.1339471159 |
- |
- |
PREDICTED: uncharacterized protein LOC104879644 [Vitis vinifera] |
| 10 |
Hb_001587_040 |
0.134407266 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000307_070 |
0.1349705811 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 12 |
Hb_002871_040 |
0.1387765446 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 13 |
Hb_000116_250 |
0.1408232169 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 14 |
Hb_001863_180 |
0.1443579078 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001307_100 |
0.1470285477 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 16 |
Hb_003529_040 |
0.1478720342 |
- |
- |
PREDICTED: ATP phosphoribosyltransferase 2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000589_170 |
0.1482416269 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_002687_160 |
0.1492685513 |
- |
- |
PREDICTED: uncharacterized protein LOC105643395 [Jatropha curcas] |
| 19 |
Hb_000230_530 |
0.1498411131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000310_060 |
0.1499173751 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |