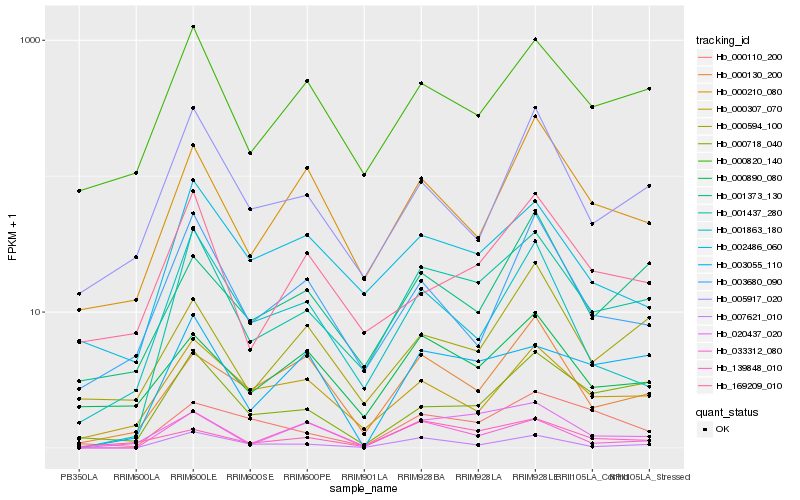
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_280 |
0.0 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 2 |
Hb_007621_010 |
0.1544315091 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000820_140 |
0.1678503475 |
- |
- |
histone H4 [Zea mays] |
| 4 |
Hb_000718_040 |
0.180987707 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 5 |
Hb_020437_020 |
0.1851748039 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 6 |
Hb_003055_110 |
0.1870265988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002486_060 |
0.1885786341 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 8 |
Hb_000307_070 |
0.1891892614 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 9 |
Hb_033312_080 |
0.1932669479 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 10 |
Hb_005917_020 |
0.1944891993 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 11 |
Hb_000210_080 |
0.1991176902 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_169209_010 |
0.2038041586 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001373_130 |
0.204830219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003680_090 |
0.2053872043 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000130_200 |
0.2066026672 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 16 |
Hb_000594_100 |
0.2092474551 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000110_200 |
0.2095008925 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000890_080 |
0.2097332107 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 19 |
Hb_001863_180 |
0.2098835514 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_139848_010 |
0.2146399945 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |