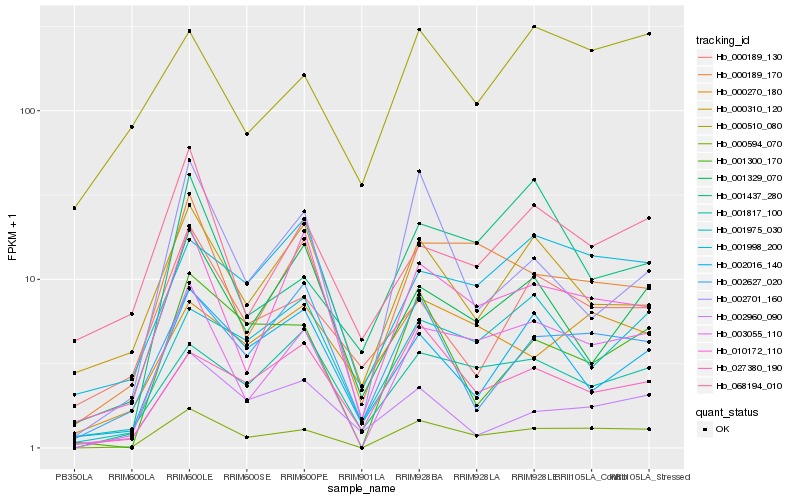
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_070 |
0.0 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_003055_110 |
0.1052597509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000189_170 |
0.1684547695 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_001300_170 |
0.1729220531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010172_110 |
0.1851828044 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 6 |
Hb_027380_190 |
0.2021381353 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002627_020 |
0.2029856041 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 8 |
Hb_001817_100 |
0.2066226059 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 9 |
Hb_002960_090 |
0.2083164914 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_002701_160 |
0.2089404404 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 11 |
Hb_000270_180 |
0.2116842958 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 12 |
Hb_000189_130 |
0.2123823709 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001998_200 |
0.2148252442 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 14 |
Hb_000310_120 |
0.2148356327 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 15 |
Hb_001975_030 |
0.2149172717 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 16 |
Hb_068194_010 |
0.2150831862 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 17 |
Hb_000510_080 |
0.21600704 |
- |
- |
hypothetical protein CICLE_v10021453mg [Citrus clementina] |
| 18 |
Hb_002016_140 |
0.2162492307 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 19 |
Hb_001329_070 |
0.2166846951 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 20 |
Hb_001437_280 |
0.2184334561 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |