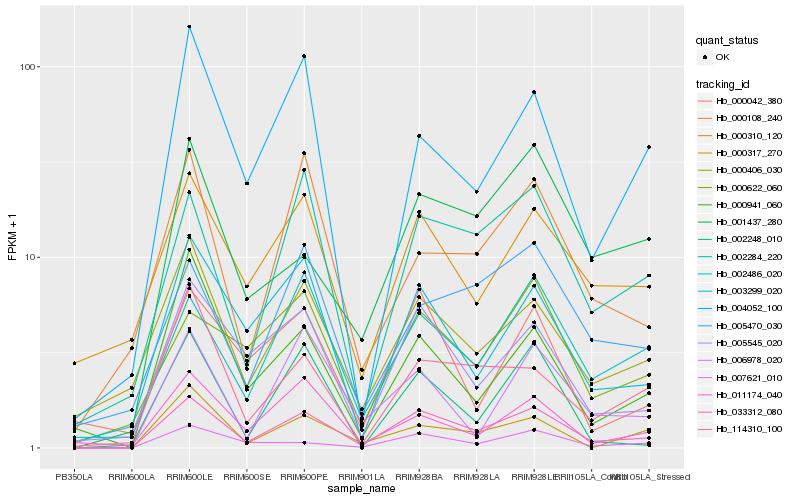
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033312_080 |
0.0 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 2 |
Hb_002284_220 |
0.1565796945 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 3 |
Hb_005545_020 |
0.1628770673 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 4 |
Hb_002486_020 |
0.1636371982 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_003299_020 |
0.1728018336 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 6 |
Hb_000406_030 |
0.1751841038 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 7 |
Hb_007621_010 |
0.1787864465 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_114310_100 |
0.1895682766 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 9 |
Hb_006978_020 |
0.1902372166 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_004052_100 |
0.1904063533 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 11 |
Hb_000317_270 |
0.1911832199 |
- |
- |
Uncharacterized protein TCM_001673 [Theobroma cacao] |
| 12 |
Hb_000310_120 |
0.1927354496 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 13 |
Hb_000108_240 |
0.1932165245 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 14 |
Hb_001437_280 |
0.1932669479 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 15 |
Hb_000622_060 |
0.1941623324 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000941_060 |
0.1949839082 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0016s05890g [Populus trichocarpa] |
| 17 |
Hb_002248_010 |
0.1957197965 |
- |
- |
PREDICTED: uncharacterized protein LOC105124180 [Populus euphratica] |
| 18 |
Hb_011174_040 |
0.1959758423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005470_030 |
0.1961699855 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000042_380 |
0.197770605 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |