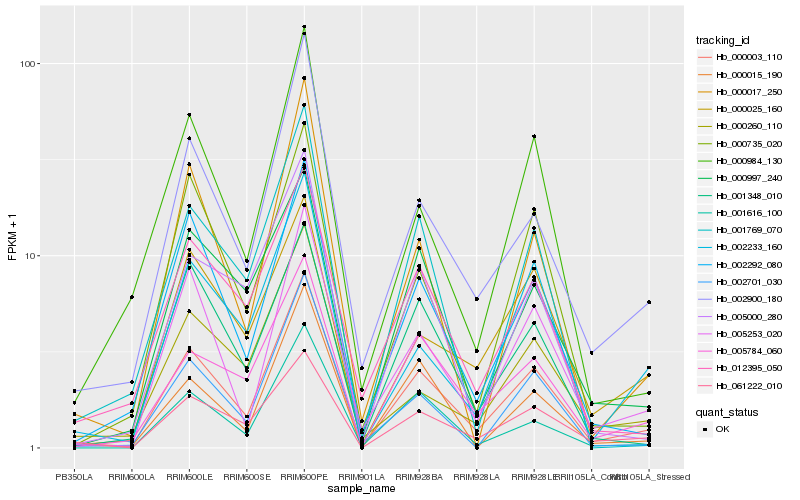
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_110 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_061222_010 |
0.1149276224 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 3 |
Hb_005784_060 |
0.1174075417 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 4 |
Hb_002900_180 |
0.1198577046 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 5 |
Hb_005253_020 |
0.1217625538 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 6 |
Hb_002292_080 |
0.124648698 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 7 |
Hb_001616_100 |
0.130587689 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 8 |
Hb_002233_160 |
0.1361792919 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 9 |
Hb_000017_250 |
0.1377811694 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 10 |
Hb_002701_030 |
0.1383431601 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 11 |
Hb_000015_190 |
0.1421607251 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 12 |
Hb_000984_130 |
0.142817269 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 13 |
Hb_005000_280 |
0.1428405248 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 14 |
Hb_000997_240 |
0.1446112472 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 15 |
Hb_000025_160 |
0.1464464665 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000735_020 |
0.146481482 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 17 |
Hb_001348_010 |
0.1468524217 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 18 |
Hb_001769_070 |
0.1471913019 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_012395_050 |
0.1490115949 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 20 |
Hb_000260_110 |
0.1493860189 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |