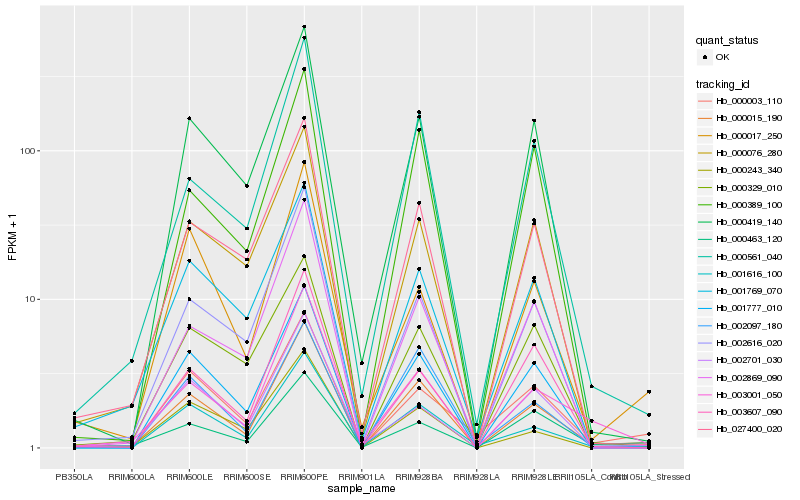
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000015_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 2 |
Hb_001616_100 |
0.111447057 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 3 |
Hb_000561_040 |
0.117610929 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 4 |
Hb_003001_050 |
0.1197006547 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 5 |
Hb_027400_020 |
0.1219838145 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001777_010 |
0.1269294415 |
- |
- |
- |
| 7 |
Hb_000419_140 |
0.1303956537 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 8 |
Hb_002616_020 |
0.1329045769 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 9 |
Hb_000076_280 |
0.1393420082 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 10 |
Hb_001769_070 |
0.1394362907 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000003_110 |
0.1421607251 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_002869_090 |
0.1424787193 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 13 |
Hb_000389_100 |
0.1432706791 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 14 |
Hb_000463_120 |
0.1498811698 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 15 |
Hb_000243_340 |
0.1499044876 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 16 |
Hb_002701_030 |
0.1502550183 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 17 |
Hb_002097_180 |
0.1522084174 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000017_250 |
0.1539557172 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 19 |
Hb_003607_090 |
0.154905097 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 20 |
Hb_000329_010 |
0.1568092744 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |