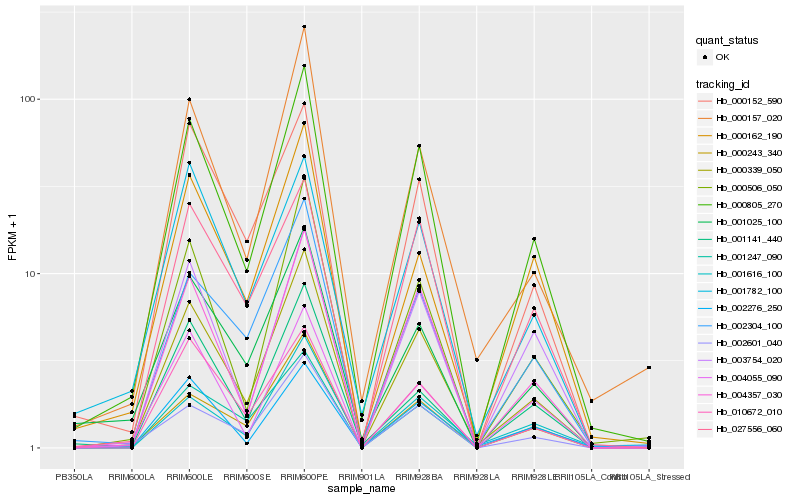
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_270 |
0.0 |
- |
- |
PREDICTED: expansin-A4 [Gossypium raimondii] |
| 2 |
Hb_000339_050 |
0.0482477729 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 3 |
Hb_004357_030 |
0.0721533647 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 4 |
Hb_000506_050 |
0.0955767958 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 5 |
Hb_002304_100 |
0.0989287095 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 6 |
Hb_002601_040 |
0.1052503028 |
- |
- |
hypothetical protein JCGZ_06197 [Jatropha curcas] |
| 7 |
Hb_000243_340 |
0.1095351011 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 8 |
Hb_003754_020 |
0.1101261062 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_001247_090 |
0.1157501356 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 10 |
Hb_002276_250 |
0.1157811628 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001025_100 |
0.1172734967 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 12 |
Hb_027556_060 |
0.1173432761 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 13 |
Hb_000152_590 |
0.1189082694 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 14 |
Hb_000157_020 |
0.1195121824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004055_090 |
0.1209596338 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 16 |
Hb_000162_190 |
0.1222912883 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 17 |
Hb_010672_010 |
0.1288754175 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 18 |
Hb_001616_100 |
0.1310080546 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 19 |
Hb_001782_100 |
0.1324123354 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_001141_440 |
0.1328723057 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |