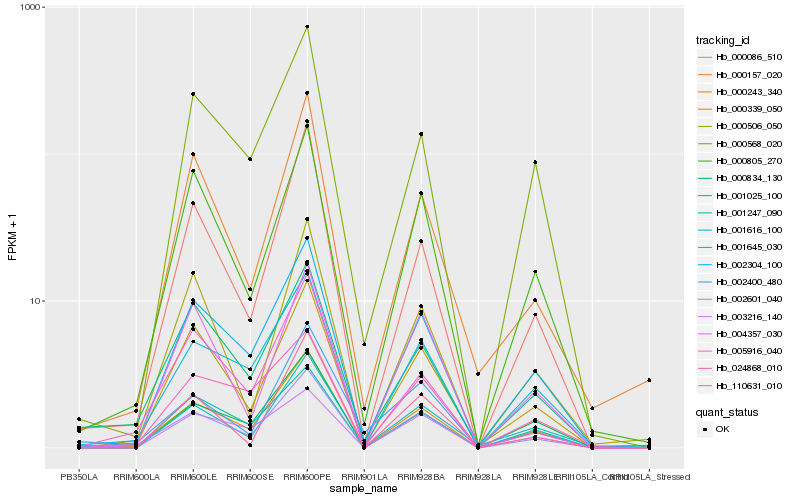
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002601_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_06197 [Jatropha curcas] |
| 2 |
Hb_000243_340 |
0.0639405919 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 3 |
Hb_002304_100 |
0.0941241191 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 4 |
Hb_000339_050 |
0.0964932693 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 5 |
Hb_001645_030 |
0.0980742237 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1b [Jatropha curcas] |
| 6 |
Hb_000805_270 |
0.1052503028 |
- |
- |
PREDICTED: expansin-A4 [Gossypium raimondii] |
| 7 |
Hb_000086_510 |
0.1285432863 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001247_090 |
0.1314590242 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 9 |
Hb_005916_040 |
0.1317703554 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 10 |
Hb_004357_030 |
0.132376293 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_001025_100 |
0.1331010107 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000834_130 |
0.1336893226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000506_050 |
0.1357898932 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 14 |
Hb_000568_020 |
0.1367166775 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 15 |
Hb_000157_020 |
0.1374965813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001616_100 |
0.1425459549 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 17 |
Hb_003216_140 |
0.1435445193 |
- |
- |
F-box protein SNE, putative [Ricinus communis] |
| 18 |
Hb_002400_480 |
0.1440534387 |
- |
- |
PREDICTED: uncharacterized protein LOC105637596 [Jatropha curcas] |
| 19 |
Hb_110631_010 |
0.1455584848 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 20 |
Hb_024868_010 |
0.1469789562 |
- |
- |
protein kinase, putative [Ricinus communis] |