| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
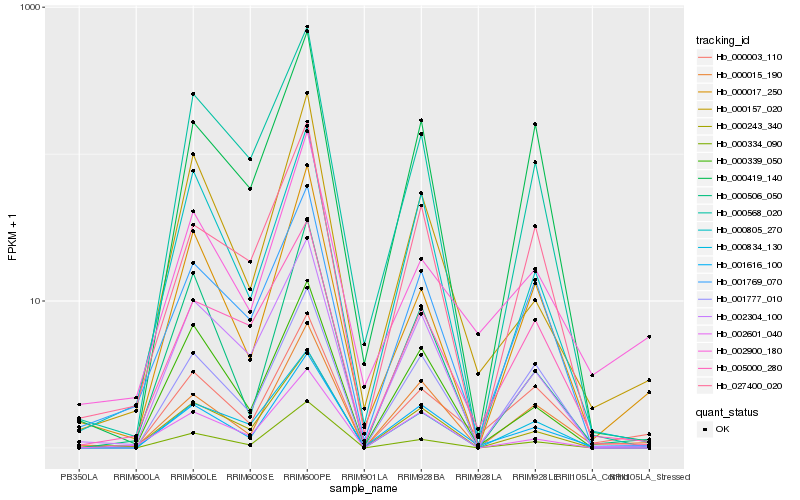
Hb_001616_100 |
0.0 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 2 |
Hb_000015_190 |
0.111447057 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 3 |
Hb_002304_100 |
0.1240660861 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 4 |
Hb_000157_020 |
0.1272062038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000243_340 |
0.1273972356 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 6 |
Hb_000003_110 |
0.130587689 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000805_270 |
0.1310080546 |
- |
- |
PREDICTED: expansin-A4 [Gossypium raimondii] |
| 8 |
Hb_000339_050 |
0.1321573854 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 9 |
Hb_000506_050 |
0.1342679432 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 10 |
Hb_001777_010 |
0.1375515123 |
- |
- |
- |
| 11 |
Hb_000419_140 |
0.1389261766 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 12 |
Hb_000834_130 |
0.1418268634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002601_040 |
0.1425459549 |
- |
- |
hypothetical protein JCGZ_06197 [Jatropha curcas] |
| 14 |
Hb_000017_250 |
0.143145371 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 15 |
Hb_000334_090 |
0.1440469694 |
- |
- |
hypothetical protein JCGZ_16789 [Jatropha curcas] |
| 16 |
Hb_000568_020 |
0.1444675197 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 17 |
Hb_005000_280 |
0.1475857639 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 18 |
Hb_002900_180 |
0.1491339679 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 19 |
Hb_001769_070 |
0.1502606891 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_027400_020 |
0.1512490444 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |