| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
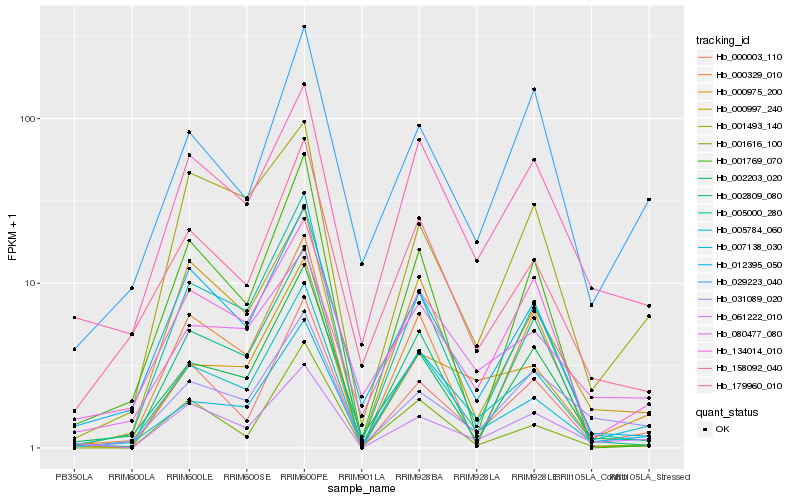
Hb_005784_060 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 2 |
Hb_007138_030 |
0.1171113139 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 3 |
Hb_000003_110 |
0.1174075417 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000975_200 |
0.1212842716 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000997_240 |
0.1252041126 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 6 |
Hb_179960_010 |
0.1262336248 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 7 |
Hb_005000_280 |
0.1302136496 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 8 |
Hb_158092_040 |
0.1302525624 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 9 |
Hb_002203_020 |
0.1313255647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_061222_010 |
0.1359913747 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 11 |
Hb_012395_050 |
0.1417669674 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 12 |
Hb_031089_020 |
0.1469003128 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 13 |
Hb_134014_010 |
0.1549673632 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 14 |
Hb_002809_080 |
0.1551550064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_080477_080 |
0.1566136519 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 16 |
Hb_029223_040 |
0.1576625402 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 17 |
Hb_001616_100 |
0.1604161555 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 18 |
Hb_001769_070 |
0.162028909 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000329_010 |
0.1623901579 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 20 |
Hb_001493_140 |
0.1637120895 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |