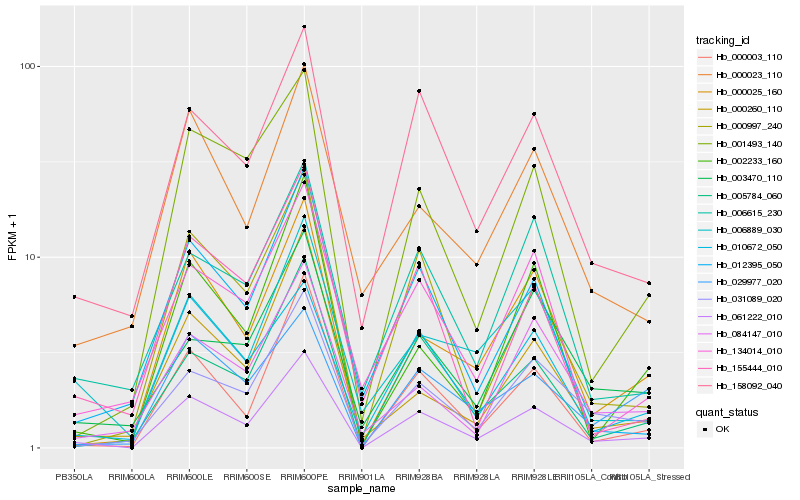
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061222_010 |
0.0 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 2 |
Hb_000003_110 |
0.1149276224 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000025_160 |
0.126685845 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 4 |
Hb_031089_020 |
0.1338017033 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_158092_040 |
0.1354558398 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 6 |
Hb_005784_060 |
0.1359913747 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 7 |
Hb_000997_240 |
0.1444063797 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 8 |
Hb_001493_140 |
0.1484357264 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |
| 9 |
Hb_002233_160 |
0.1534265995 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 10 |
Hb_010672_050 |
0.1564643207 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 11 |
Hb_006615_230 |
0.1581543456 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 12 |
Hb_006889_030 |
0.1617199752 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 13 |
Hb_012395_050 |
0.1618190865 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 14 |
Hb_134014_010 |
0.1623831526 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 15 |
Hb_000260_110 |
0.16255195 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 16 |
Hb_029977_020 |
0.1634237167 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000023_110 |
0.1647088922 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 18 |
Hb_084147_010 |
0.1655075483 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_155444_010 |
0.1663257555 |
- |
- |
- |
| 20 |
Hb_003470_110 |
0.1665344358 |
- |
- |
unnamed protein product [Vitis vinifera] |