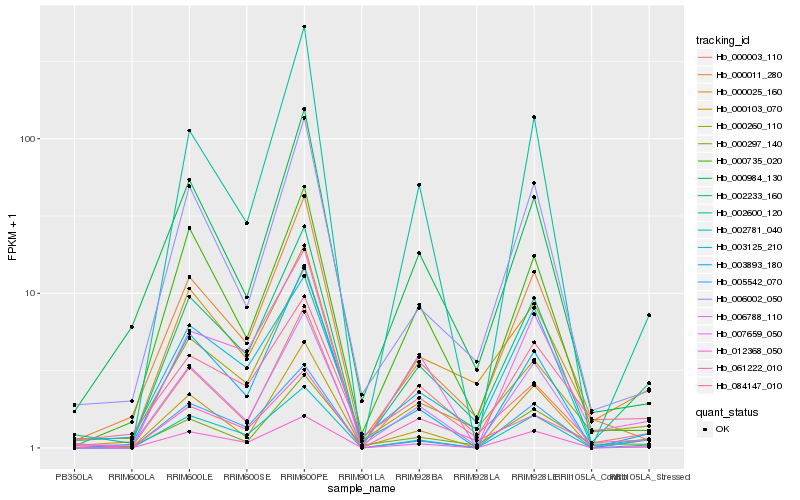
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002233_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 2 |
Hb_000260_110 |
0.1115234298 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 3 |
Hb_005542_070 |
0.1184768957 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 4 |
Hb_012368_050 |
0.118884605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002781_040 |
0.1196184742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000025_160 |
0.1231580184 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006002_050 |
0.1313965503 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 8 |
Hb_000103_070 |
0.1322669798 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 9 |
Hb_003893_180 |
0.1323574748 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 10 |
Hb_000011_280 |
0.1330067846 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 11 |
Hb_000003_110 |
0.1361792919 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_007659_050 |
0.1404419572 |
- |
- |
PREDICTED: xylogen-like protein 11 [Jatropha curcas] |
| 13 |
Hb_000297_140 |
0.1416978147 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 14 |
Hb_006788_110 |
0.1433412226 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 15 |
Hb_084147_010 |
0.1455817529 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_003125_210 |
0.1474982064 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 17 |
Hb_000984_130 |
0.1485687873 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 18 |
Hb_000735_020 |
0.1507987959 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 19 |
Hb_002600_120 |
0.1514222449 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 20 |
Hb_061222_010 |
0.1534265995 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |