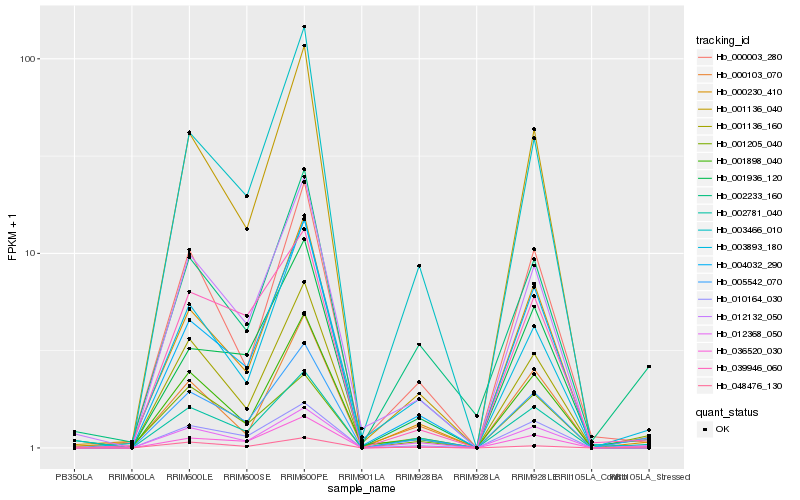
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005542_070 |
0.0 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 2 |
Hb_002781_040 |
0.0599375093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012368_050 |
0.0998885612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002233_160 |
0.1184768957 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 5 |
Hb_003893_180 |
0.1205238156 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 6 |
Hb_000230_410 |
0.1241501831 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 7 |
Hb_000103_070 |
0.1251587883 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 8 |
Hb_012132_050 |
0.1303176261 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 9 |
Hb_010164_030 |
0.1319751006 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_036520_030 |
0.1341701122 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 11 |
Hb_003466_010 |
0.1354075091 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 12 |
Hb_001136_160 |
0.1360771971 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_039946_060 |
0.1374583293 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 14 |
Hb_004032_290 |
0.1377669895 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_048476_130 |
0.137906119 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 16 |
Hb_000003_280 |
0.1380175042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001936_120 |
0.1382598615 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001205_040 |
0.1391121534 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_001136_040 |
0.1399055083 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_001898_040 |
0.1399504631 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |