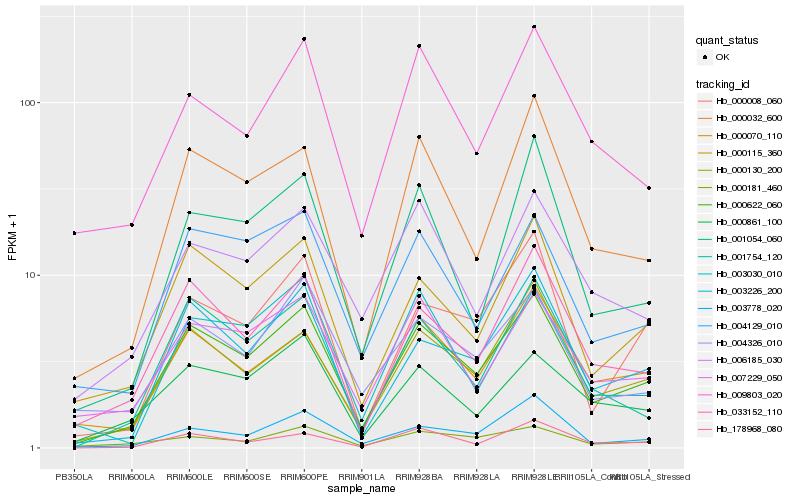
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000622_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000115_360 |
0.0936318372 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000130_200 |
0.0937552904 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 4 |
Hb_033152_110 |
0.1007210949 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 5 |
Hb_003226_200 |
0.1030123155 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 6 |
Hb_000032_600 |
0.1094622234 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 7 |
Hb_004129_010 |
0.1148975884 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_001754_120 |
0.1149577862 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 9 |
Hb_000070_110 |
0.1157075669 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007229_050 |
0.1167710052 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004326_010 |
0.1244516366 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_003778_020 |
0.1261561983 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_000861_100 |
0.1266899 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000008_060 |
0.1274565986 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_178968_080 |
0.1287131885 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_006185_030 |
0.1308351755 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 17 |
Hb_001054_060 |
0.1314942663 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 18 |
Hb_009803_020 |
0.1317694304 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 19 |
Hb_000181_460 |
0.1328420284 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 20 |
Hb_003030_010 |
0.1349343706 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |