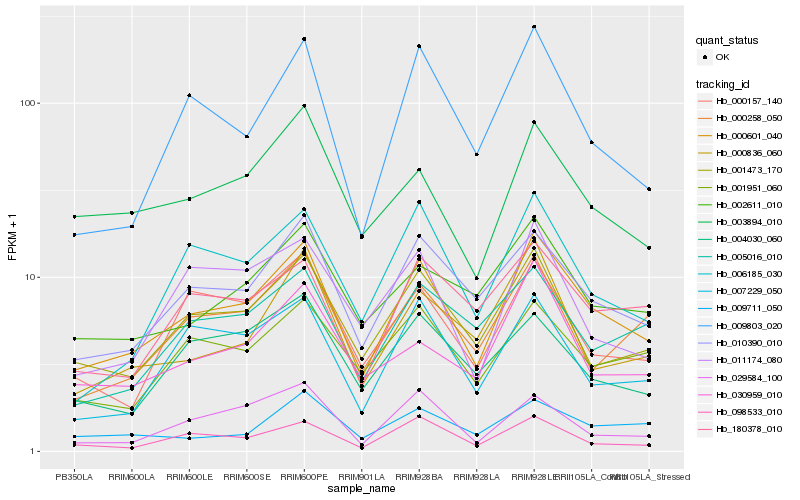
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000601_040 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 2 |
Hb_010390_010 |
0.1030761042 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 3 |
Hb_001473_170 |
0.10907535 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 4 |
Hb_009803_020 |
0.1177353079 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 5 |
Hb_098533_010 |
0.1223216092 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 6 |
Hb_006185_030 |
0.1254544849 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 7 |
Hb_001951_060 |
0.1260624876 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 8 |
Hb_003894_010 |
0.1267659583 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_007229_050 |
0.1274690965 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002611_010 |
0.1297957738 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000258_050 |
0.1298950015 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_180378_010 |
0.130580948 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 13 |
Hb_000157_140 |
0.1327712974 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_011174_080 |
0.1358009521 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 15 |
Hb_030959_010 |
0.1366055052 |
- |
- |
cullin, putative [Ricinus communis] |
| 16 |
Hb_000836_060 |
0.1370613334 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 17 |
Hb_005016_010 |
0.1376410531 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 18 |
Hb_029584_100 |
0.1394678117 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 19 |
Hb_009711_050 |
0.1397053476 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 20 |
Hb_004030_060 |
0.1398505395 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |